Reactivity in Chemistry
Enzyme Catalysis
EZ1. Introduction to Enzymes
If you look around the laundry room cupboard, you might see some different household products that contain enzymes. That might seem a little strange at first; enzymes are found in the body, and have no business being in stain remover or drain cleaner. But enzymes are just molecules that are really, really good at performing specific tasks. In these household applications, enzymes are really good at breaking specific bonds and replacing them with new ones. In doing so, they are breaking down larger molecules, which might be causing a stain or a slow drain. Those bigger molecules turn into smaller molecules that can be washed away more easily.
What kind of molecules are enzymes? They are special proteins. Proteins play lots of roles in biology; they may provide structure, for example, or they might catalyse reactions. Enzymes are proteins that catalyse reactions. Proteins belong to a class of molecules called biopoylmers. Biopolymers are large molecules composed of smaller molecules that have been bonded together. The small building blocks of proteins are the amino acids. One of the important features of amino acids is the fact that they are chiral; they have very specific three-dimensional shapes, like a left-handed glove or a right-handed glove.
Enzymes are proteins
Proteins are composed of chiral amino acids
Enzymes are great, big, chiral molecules
How big are these proteins, exactly? The biggest protein is titin, which is partly responsible for the elastic properties of muscle. Titin is composed of about 35,000 individual amino acids. The molecular weight of an amino acid is roughly 100 Da (100 Daltons or 100 amu), so that means titin has a molecular weight of about 35 million Da.
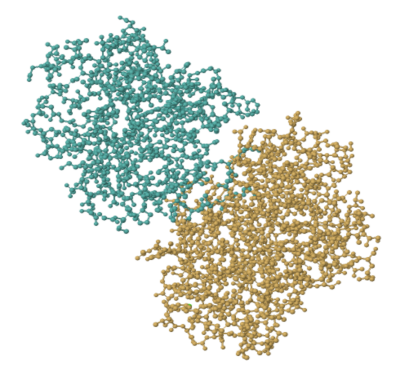
But titin is one of those structural proteins. Let's look at an enzyme. Trypsin is a really common enzyme, found in the gut. The picture below comes from a structure determination experiment involving pig trypsin. It's made of about 450 amino acids, so its molecular weight is about 45,000 Da.

FigureEZ1.1. The experimentally observed structure of trypsin.1
That picture shows every atom other than the hydrogens in trypsin; the hydrogens are too small to bother with. Even so, it's a bit busy. More often we look at enzymes and other proteins in a simplified, cartoon form. The cartoon model of trypsin below highlights the beta sheets and alpha helices, the secondary structures within trypsin. In the cartoon model, you can get a better sense of the shape of things. You can see gaps and grooves in the protein, and you can see the chirality in those helices.
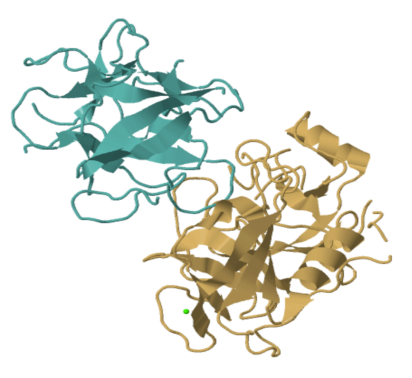
FigureEZ1.2. The structure of trypsin presented in "cartoon" form, showing sheets, loops and helices instead of atoms.1
Trypsin belongs to a class of enzymes called hydrolases. Hydrolases break bonds with the help of a water molecule. Trypsin breaks specific amide or peptide bonds in other proteins (yes, it's a cannibal), converting the carbonyl side of the bond into a carboxylic acid. That starts breaking down proteins in food; it's an essential part of digestion.
But how does trypsin do that?
At the most basic level, enzymes bind their targets, called substrates, perform chemical reactions on them, and let them go. Binding the substrate is a key step, and it's one of the reasons the chirality of enzymes can be important. Some enzymes are extremely specific in what substrates they bind. For example, they might bind one enantiomer and not another.
Trypsin is also very specific; it specializes in breaking the amide bond next to either arginine or lysine residues. The reaction that it catalyses is not necessarily specific to that particular peptide bond, but the trypsin selectively binds at that site.
This role of specific binding in enzymes is often referred to as the "lock and key" mechanism. We think of enzymes as being unlocked or turned on when the proper key is inserted, causing the enzyme to spring into action. In the following sections, we'll take a closer look at how enzymes bind their substrates and carry out their reactions.
Problem EZ1.1.
Enzymes often have two-part names: the first part identifies the substrate, and the second part describes what the enzyme does with that substrate. See if you can tell what an enzyme would do if one of the following words was in its name:
Match each item in one column to an item in the other column.
a) isomerase i) add oxygen from O2 into a molecule
b) hydrolase ii) add both oxygens from O2 into a molecule
c) oxygenase iii) oxidize or remove electrons from the substrate
d) dioxygenase iv) reduce or add electrons to the substrate
e) reductase v) reorganize atoms from one isomer into another
f) transferase vi) cleave a phosphate group off a protein
g) phosphatase vii) add water into a molecule, helping to break it down
h) oxidase viii) cause two molecules to be bound together
i) ligase ix) transfer a functional group to or from a molecule
1. Trypsin Structure: PDB ID: 4AN7
Patil, D.N., Chaudhary, A., Sharma, A.K., Tomar, S., Kumar, P. Structural Basis for Dual Inhibitory Role of Tamarind Kunitz Inhibitor (Tki) Against Factor Xa and Trypsin. FEBS J. 2012, 279, 4547.
This site was written by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with other authors as noted on individual pages. It is freely available for educational use.

Structure & Reactivity in Organic,
Biological and Inorganic Chemistry by
Chris Schaller
is licensed under a
Creative Commons Attribution-NonCommercial 3.0 Unported License.
Send corrections to cschaller@csbsju.edu
This material is based upon work supported by the National Science Foundation under Grant No. 1043566.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Navigation: