Reactivity in Chemistry
Photosynthesis
PS4. Photosystem II: The Oxygen-Evolving Complex
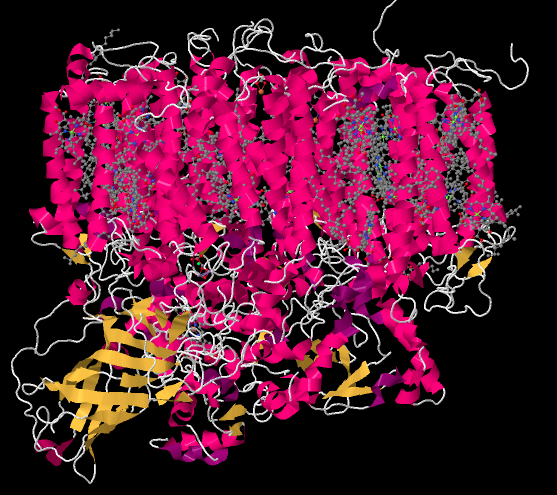
You may recall that the oxygen-evolving complex is found deep in Photosystem II. It's found near that yellow-coded bundle of beta-sheets in the lower left of the picture below; that part of the complex is called the manganese-stabilizing protein. Remember, Photosystem II is embedded in the thylakoid membrane. The membrane surrounds the pink portion of alpha-helices in the top half of the picture. The stroma, the aqueous medium that surrounds the thylakoid, is at the very top of the picture. In the lower part of the picture, the protein is dipping its toes into the lumen; that's the watery medium inside the thylakoid.

Figure PS4.1. X-ray crystallographic structure of photosystem II in the marine cyanobacterium Thermosynechococcus elongatus.1
When we strip away all of the proteins, we are left with a number of ligands that were embedded inside. That includes the light-harvesting complex that we have already looked at, as well as the small clutch of atoms called the oxygen-evolving complex (OEC). The OEC is found a little further down in the picture, at the edge of the lumen. You can see its manganese atoms, coloured purple in this picture, as well as red oxygens and grey calcium, mid-way down and a little to the left.
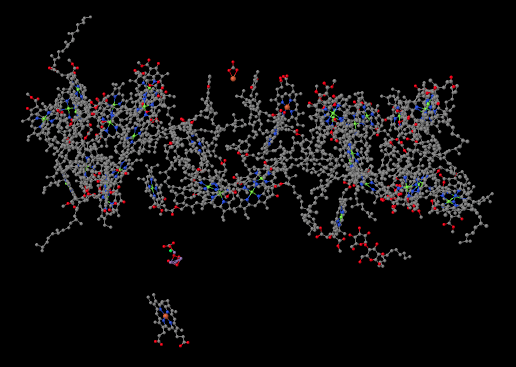
Figure PS4.1. X-ray crystallographic structure of photosystem II, showing only the ligands.1
Let's zoom in for a closer look. A couple of stray atoms are visible in the picture, but focus on the atoms that are connected together. The "spokes" mean that the X-ray crystallography software has decided these atoms are close enough together that they are bonded to each other. The purple manganese atoms might not really be bonded to each other in terms of sharing a pair of electrons between the atoms, but they are magnetically coupled, sharing electrons in a different sense. We can see that the atoms almost form a cube, although it has one corner torn open. It's not unlike an FeS cluster, only with manganese and oxygen instead of iron and sulfur.
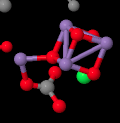
Figure PS4.3. X-ray crystallographic structure of photosystem II: a close-up of the oxygen-evolving complex (in a different orientation from the figure above).1
The task of this little cube is to carry out a reaction that is indispensible for our existence on earth. It needs to convert water into oxygen. From an evolutionary point of view, there is a distinct advantage to the use of water as a source of electrons. There is an almost endless supply: we have oceans full of it on this planet. The trouble is, removing an electron from water results in an oxygen radical; in biochemistry, this is called a reactive oxygen species. Reactive oxygen species can be chaotic, evil characters inside a cell, reacting indiscriminately with any other structures that they contact. For that reason, it would be safer to oxidise the water all the way to dioxygen.
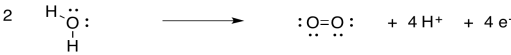
Scheme PS4.1. The oxidation of water to dioxygen.
Problem PS4.1.
Use the Lewis structures to confirm the number of electrons involved in the oxidation of water.
That four-electron oxidation of water is a pretty challenging task. The opposite event, the four-electron reduction of dioxygen to water, served as the thermodynamic sink for the electron transport chain in oxidative phosphorylation. Nevertheless, it was also pretty challenging, because it required the rapid introduction of four electrons into the system in order to fully reduce the dioxygen to water, without the formation of free reactive oxygen species that might wreak havoc in the cell.
In that case, the problem was solved through a large number of iron and copper atoms within Complex IV, serving as a reservoir of electrons for rapid reduction. Furthermore, the actual site of reduction was a close-set iron and copper pair, allowing the dioxygen to bridge between two metals to aid in that rapid reduction.
The oxygen-evolving complex uses a similar strategy, but with a reverse bias. The cluster of manganes ions provides a reservoir of holes rather than electrons, allowing for the rapid oxidation of water to dioxygen. The role of the calcium ion is not clear, since calcium is not redox-active; it is always Ca2+. Remember, metals in the first few columns of the periodic table are always found in their maximum oxidations states, corresponding to a noble gas configuration. However, it may play another role in binding water or stabilizing the cluster.
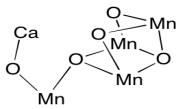
Figure PS4.4. The oxygen-evolving complex (cluster only; coordinating amino acid residues neglected).
Problem PS4.2.
It can be useful to know which metal ions are typically redox-active and which are not (i.e. those metals that are always found in the same oxidation state). For the following metals, indicate whether the metal is redox-active and provide its most common oxidation state(s).
a) Fe b) Mg c) Cu d) Li
e) Zn f) Sc g) Co h) Cr
Remember that there is an intermediary between P680+* and the oxygen-evolving complex. There is a tyrosine residue, called Tyr161 because of its position in the primary structure of the protein, situated between the oxygen-evolving complex and P680+*. It conducts the electron from manganese to the chlorophyll in the reaction centre. An electron is first transferred from Tyr161 to P680+*. An electron from manganese then replaces the missing electron on Tyr161.
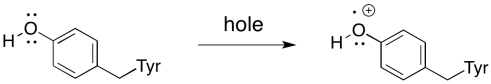
Scheme PS4.2. Oxidation of tyrosine 161.
It takes the removal of four electrons to oxidise water to oxygen, so all of that has to happen four times. Four photons must be absorbed, exciting P680 four times. Each time, an electron from Tyr161 replaces the electron that was excited from the ground state in P680, leaving behind a hole. The electron in Tyr161 is replaced by an electron from manganese. These events are illustrated in an overall mechanism called the Kok cycle (pronounced "coke" cycle).
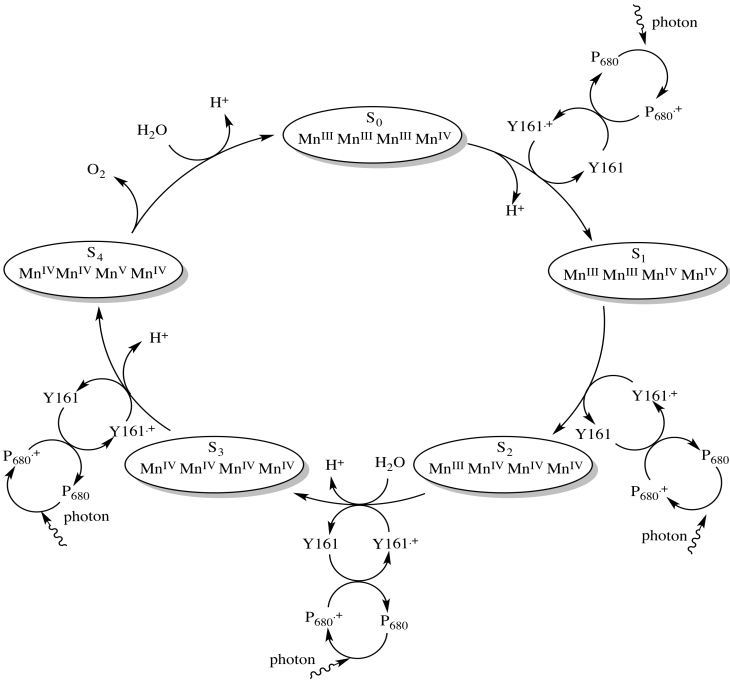
Scheme PS4.3. The Kok cycle, relating photon absorption, manganese oxidation, and water oxidation.
The Kok cycle illustrates the storage of holes on the manganese atoms. The holes are just the places where electrons used to be. We can think of them as a buildup of positive charge. The cycle also shows one of the steps in water oxidation that is evident in the equation for the reaction: the loss of four protons. The loss of protons is probably linked directly to the oxidation of the manganese ions. Any time an atom or molecule becomes more positively charged, it becomes more highly acidic.
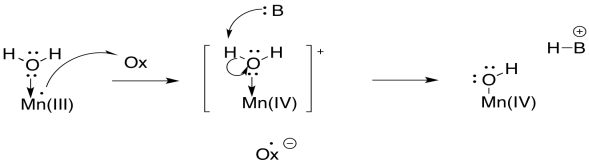
Scheme PS4.4. Deprotonation of a water molecule linked to manganese oxidation.
From the point of view of the water molecule, just binding to a manganese makes it more likely to lose a proton, because it is donating electrons to another atom and thereby taking on a formal positive charge on oxygen. So, a water molecule becomes much more acidic when it binds to a metal atom. That's a common event in biochemistry. The buildup of additional positive charge on manganese makes this deprotonation even more likely.
In terms of how the O-O bond is formed, different pathways are conceivable, but the simplest possibility involves activation of the water as a nucleophile. Deprotonation results in a hydroxide ion. Although the hydroxide is bound to the metal atom, the polarity of the bond allows the hydroxide to behave as a nucleophile.
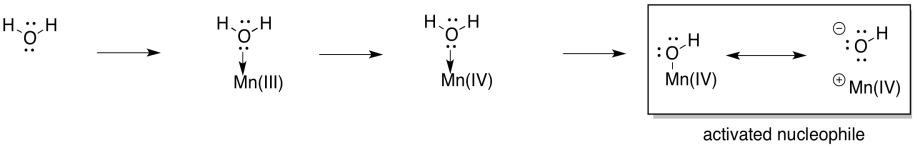
Scheme PS4.5. Metal binding and deprotonation activates water as a nucleophile.
At this point, you have also seen water become activated as a nucleophilic hydroxide ion in other biochemical pathways, so this development should not be a big surprise. What about an electrophile? Well, one way to provide an electrophile is to deprotonate the hydroxide to make a metal oxo. That deprotonation could be linked to another oxidation at manganese.
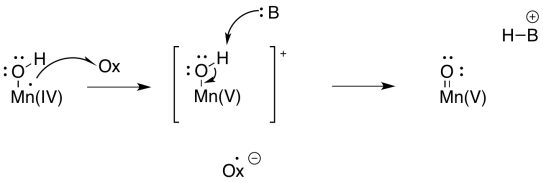
Scheme PS4.6. Deprotonation of a hydroxy ligand, forming a metal oxo complex, linked to manganese oxidation.
Metal oxo compounds can develop electrophilic character at oxygen because of π-donation from oxygen to a metal ion in a high oxidation state. The donation of a lone pair to the metal would give oxygen a formal positive charge. The oxygen would be activated as an electrophile.
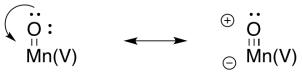
Scheme PS4.7. π-Donation from oxygen in a metal oxo complex, showing electrophilic character on oxygen.
At that point, it is easy to imagine how one oxygen (a hydroxy ligand) could act as a nucleophile while a second (an oxo ligand) acts as an electrophile. An O-O bond would form, accompanied by a two-electron reduction at manganese.
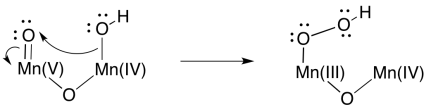
Scheme PS4.8. A possible O-O bond-forming event, accompanied by manganese reduction.
Formation of a second O=O bond would result in a second two-electron reduction at manganese. In two easy steps, the entire four-electron reduction of manganese -- and four-electron oxidation of water -- has been accomplished. Because there are four manganese ions within the cluster, it is a simple matter to redistribute electrons so that the manganese ions have similar oxidation states.
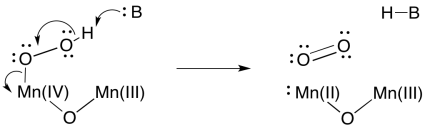
Scheme PS4.9. A possible O=O bond-forming event, accompanied by manganese reduction.
Of course, because we are dealing with a manganese oxo cluster rather than an individual manganese ion, electrons can be reshuffled at any time. Oxidation states in clusters are complicated, because of the ease of transferring or even delocalising electrons within the system.
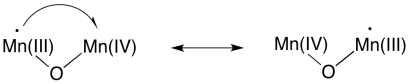
Scheme PS4.10. Electronic resorganisation within the manganese oxo cluster.
Again, there are other possible mechanisms, and you know enough chemistry at this point that you should be able to propose some alternatives.
Problem PS4.3.
a) Propose an alternative mechanism for oxygen evolution using classic organometallic mechanistic steps (oxidative addition, reductive elimination, insertion, beta-elimination).
b) What side products might be detected if this mechanism operated?
Problem PS4.4.
a) Propose an alternative mechanism for oxygen evolution using radical mechanistic steps.
b) What side products might be detected if this mechanism operated?
One last point about the oxygen-evolving complex. The goal of photosynthesis is to make ATP; that ATP will be used to convert CO2 to carbohydrates. The way to make ATP in this case is to use an electron transport chain to release enough energy to generate a proton gradient. The water is the starting point for that electron transport chain. But wait, there is a bonus: just by oxidising the water, we have released protons into the lumen. That's a head start on setting up a proton gradient.
1. X-ray crystal structures: Ferreira, K.N.; Iverson, T.M.; Maghlaoui, K.;
Barber, J.; Iwata, S. Architecture of the photosynthetic oxygen-evolving
center. Science 2004, 303, 1831-1838. Images
obtained via RCSB Protein Data Bank (1S5L).
This site is written and maintained by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with contributions from other authors as noted. It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
This material is based upon work supported by the National Science Foundation under Grant No. 1043566.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Navigation: