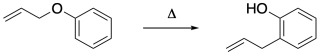
Determination of Mechanism in Chemistry
Detection of Intermediates
DI1. Isotopic Labeling
We know that a given element has a constant number of protons in its nucleus. We also know that some atoms of that element can have different numbers of neutrons in their nuclei. The number of nuclei has very little effect on the behaviour of the atom; 13C and 12C both form four bonds and they form all of the same compounds, but the molecules containing 13C are a little heavier than the ones containing the 12C.
Obviously, because they have different masses, a 12C atom and a 13C atom show up at different positions in a mass spectrum, and so do the compounds containing those atoms. In addition, a 13C nucleus can absorb radio waves when placed in a magnetic field -- it is "NMR-active" -- whereas a 12C atom cannot. Furthermore, a 14C nucleus is radioactive, opening up avenues to detect its presence with a scintillation counter. So, there are ways of detecting whether a given atom in a specific position within a molecule is a 12C or a 13C.
We can find other examples of atoms that have different isotopes that can be detected in different ways. The most common isotope of hydrogen is protium, 1H, which is NMR-active. It may come as a surprise that deuterium, 2H, is also NMR-active -- surprising because of the fact that we often use deuterated solvents when we are performing NMR spectroscopy. Typically, we are using deuterated solvents so that the hydrogens in the solvent don't overwhelm the signal of the solvent, because deuterium doesn't show up in the 1H NMR spectrum. It does, however, show up in the 2H NMR spectrum; it just absorbs different frequencies of radio waves than protium, so their spectra do not overlap. In fact, you have probably noticed coupling to deuterium in a carbon NMR spectrum; if you use CDCl3 as solvent, the carbon attached to deuterium shows up as a 1:1:1 triplet. Finally, tritium, or 3H, is radioactive and consequently it can be detected with appropriate equipment.
As a result, if we suspect a specific bonding change occurs during the course of a reaction, we can confirm the idea through an isotopic labelling experiment. Suppose we place a specific isotope of an atom in a certain position within a molecule. At the end of the reaction, is it found here, or has it moved over there?
The Claisen rearrangement is a classic example that is sometimes used to illustrate this idea. In this allyl phenyl ether variation, the reaction involves movement of the allyl group from the oxygen to an ortho- carbon.
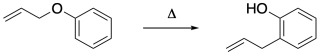
How does that change occur? Given the relative stability of both the phenol anion and the allyl cation, it doesn't seem to be too much of a stretch to propose that this is a polar reaction involving charged intermediates. Maybe a C-O bond ionizes and the resulting phenolate anion reconnects with the allyl cation through its pseudo-enolate ortho- position.
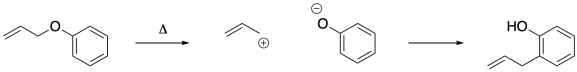
That sequence implies an intermolecular reaction. At some point, there are two molecules (two ions, really) coming together, even though there was just one molecule in the beginning and one molecule in the end. We can tell whether that is happening through something called a double labeling crossover experiment. We can label two different parts of the molecule and see whether they are still in the same molecule at the end of the reaction. There are different ways to do that experiment: we could start with one doubly-labeled compound and one non-labeled compound or, as shown below, start with one compound in which one site is labeled and another compound in which the other site is labeled.
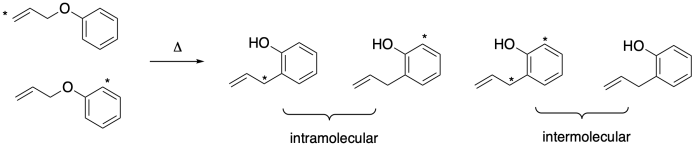
If the reaction is intermolecular, then at some point we would have four separate pieces available for partnering: labeled and non-labeled allyl as well as labeled and non-labeled phenol. That leads to four different combinations or four different isotopomers. If the reaction is intramolecular, we still have only a singly-labeled molecule at the end. If the reaction is intermolecular, then in addition to the isotopomers that would result from the intramolecular pathway, we also get a doubly-labeled compound and a non-labeled compound. We could easily distinguish these products in a mass spectrometer, because the intermolecular reaction produces two additional molecular ions.
For example, if we started with deuterium labels at the end of the allyl double bond (i.e. CD2=C) and in the two ortho positions of the phenol, the molecular ion of both the reactant and the product would show up at m/z = 134 if the reaction were intramolecular. If it were intermolecular, we would also see product molecular ions at M+-2 and M++2, or at m/z = 132 and 136. If you run an experiment like this, the reaction appears to be entirely intramolecular. There are no ions.
If we consider how that intramolecular reaction could happen (and you may already know the answer), there are a couple of ways that, on paper, at least, you could draw the arrows. Either way we do it, we are going to end up invoking a keto-enol tautomerism step at the end. Prior to that, we can picture the allyl shifting over to the ortho- position in a four-electron step, or reaching down to form a six-membered ring transition state in a six-electron step.
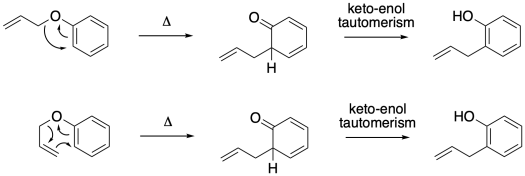
This time, a simple isotopic labeling experiment can distinguish the two possibilities. If we can label the allyl group at one end or the other, we can distinguish which end attaches to the benzene. We would find that the terminal end of the alkene moves into the benzylic position in the product because of the thermally allowed pericyclic transition state.
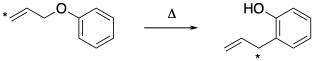
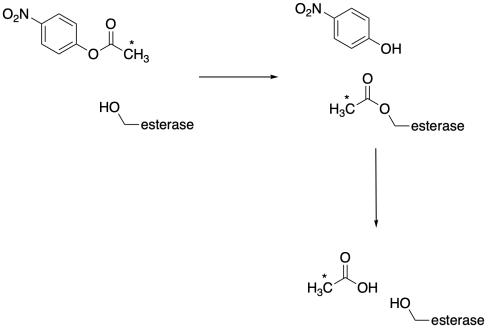
Isotopic labeling is invaluable in biochemistry because the relatively complicated molecular environment and very small scale of reactants makes biochemical reactions difficult to follow by a number of other methods. For example, researchers have made significant use of these tools in establishing the role of group transfer in enzymes capable of performing ester hydrolysis. Consider what happens if we introduce a label into the acetyl group in the acetyl ester of p-nitrophenol. This ester is often used to study esterases because it is a chromogen, producing a brightly-coloured p-ntrophenol upon hydrolysis that can be monitored conveniently by UV-Vis spectroscopy or fluorimetry, even at very low concentrations. In order to monitor that acetyl group, we would probably us 14C, because very low levels of this radioactive isotope can be detected by a scintillation counter or other device for monitoring radioactivity.

Group transfer is a common enzymatic strategy in cases like this. How do we know that? Because if we slow down the reaction (by changing pH, for example), we can actually catch the radio-labeled acyl group attached to the enzyme. If we then adjust the pH to optimum levels for the enzyme, the reaction proceeds again. That part is important because it tells us that this acylenzyme intermediate is "kinetically competent". It isn't some dead-end side product that coincidentally formed when we messed with the reaction conditions; it is capable of turning over and allowing the enzyme to continue catalyzing the reaction.
Alternatively, there are lots of other situations in which isotopic labels are useful in biological chemistry. Suppose we want to establish whether a specific compound plays a role in a reaction pathway. This consideration may be especially vexing in biochemical pathways, because we don't have absolute control over the chemical makeup of a cell. Consequently, a fragment introduced into a compound at some point might come from source A, or it might come from source B. By labeling that source with an isotope, then looking for the isotope to show up in the ultimate product, we may get a better idea about what is going on. This approach is often refered to as the use of an isotopic tracer. We feed in the isotopes and trace where they come out, learning something about what has happened in between.
Problem DI1.1.
The oligomerization of alkenes is economically important in the synthesis of compounds containing long alkyl chains, such as soaps. Researchers at Caltech used a mixture of C2H4 and C2D4 to investigate the mechanism of chromium-catalyzed hexene formation. There are two proposed mechanisms:
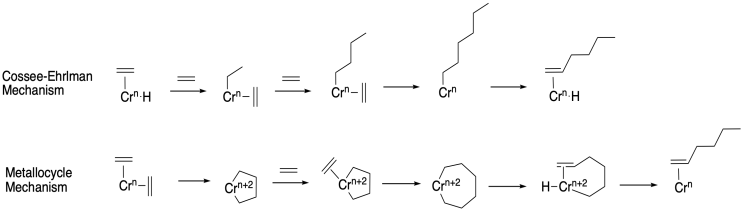
The reaction gave a mixture of C6H12: C6H8D4: C6H4D8 : C6D12: . Which mechanism is consistent with this observation?
This site is written and maintained by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (with contributions from other authors as noted). It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
Send corrections to cschaller@csbsju.edu
Navigation:
Back to Determination of Mechanism
Back to Structure & Reactivity