Determination of Mechanism in Chemistry
Detection of Intermediates
DI5. Site-Directed Mutagenesis
In biochemistry, enzymes play a central role in carrying out chemical reactions. Enzymes are catalysts, so they carry out transformations through alternative mechanistic pathways. The side chains or amino acid residues in these proteins play crucial roles in binding substrates and promoting subsequent reactions. Understanding what role is played by a specific amino acid in the protein is crucial to understanding how a biochemical reaction occurs.
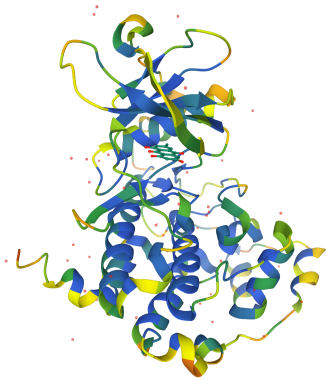
These days, thoughts about which amino acids play critical roles in an enzyme-catalyzed reaction are often guided by X-ray crystal structures of the proteins in question. Often, the active site in that structure can be identified based on comparison with other enzymes. Sometimes, a competitive inhibitor can be bound in the enzyme active site, further highlighting the area. The example below is a phosphatase with a competitive inhibitor (in grey).

If we think we know something about the active site of the enzyme, the area in which the substrate is bound in order to undergo a subsequent reaction, we can probe the role of specific amino acids by replacing them with alternative residues. Maybe we think a glutamate is well-positioned to remove a proton from the substrate. If we change the glutamate to an alanine, what happens to the reaction? Does it still work just as well? If not, maybe we have identified a crucial part of the machinery. If there isn’t much change, we can move on and look at another site.
Problem DI5.1.
In addition to playing roles in acid-base steps, amino acids may also act as ligands for metal cofactors. In each of the following pairs, indicate which amino acid would be likely to act as a ligand for a metal ion.
a) Cys or Ala b) Val or Asp c) Trp or His
Problem DI5.2.
Mutations can also be used to probe sterics in the active site. Indicate whether the following changes to the active site would result in a larger or smaller binding pocket.
a) A134L b) V256G
How do we change just one amino acid residue in a protein? First we need to know where the residue is located in the primary structure of the protein, which is just the order in which the amino acids are connected together. Maybe it is a histidine and it is the 137th amino acid in the peptide chain. Of course, the amino acid has two ends, so we had better all be in agreement about which end we start counting from. We always count from the amino end or N-terminus towards the carboxylate end or C-terminus.
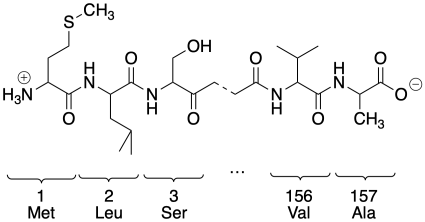
The original protein consists of a series of amino acids enchained in a particular order. This one has a valine at the 156th position. If we were to switch out that valine for a glycine, we would get a mutant protein. We would call it V156G, meaning we had switched out the 156th amino acid, a valine, and replaced it with a glycine. The original protein is referred to as the wild-type protein, or WT. It's what the protein looks like in its wild, natural state.
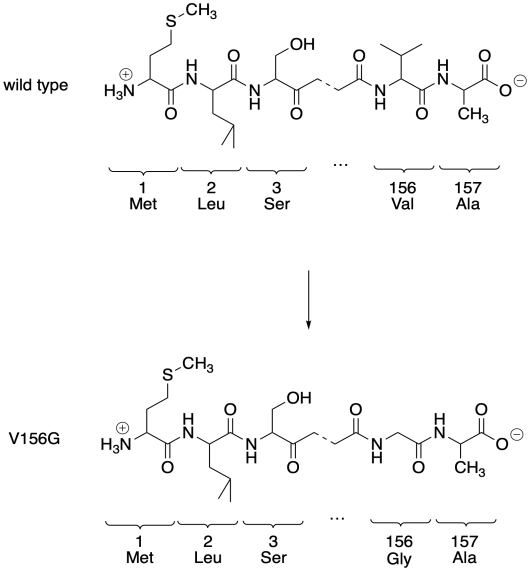
We can control the identity of any single amino acid in that polypeptide chain using RNA. The cellular machinery that assembles proteins, the ribosomes, read mRNA sequences and translate them into proteins. A sequence of three consecutive RNA bases signifies one particular amino acid residue that should be enchained into the polypeptide sequance. Changing a set of these bases is all that's needed to instruct a cell to manufacture a protein with a substituted amino acid.
We can look at a codon table to see what base pairs in mRNA would be translated into a particular amino acid. For example, the mRNA sequence UUU would signal for a phenylalanine (Phe, F) to be incorporated in the protein chain. The ribosome would similarly incorporate other amino acids into the protein in the order that their codons were encountered. Eventually, a stop codon such as UGA informs the ribosome that the protein sequence has been completed.
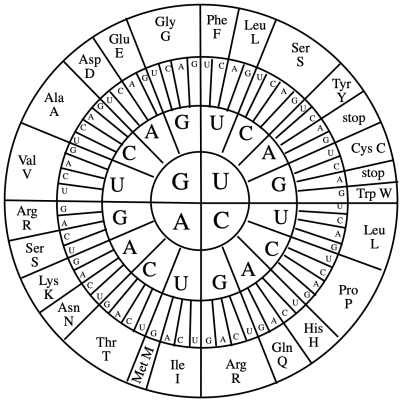
Problem DI5.3.
Provide the amino acid sequence in the peptide coded for in each of the following mRNA sequences.
a) AUGAAGGUCUAA
b) AUGUGGGAGUAG
c) AUGUGCAGAUGA
The pictures below are from X-ray structures of wild-type carbonic anhydrase and a mutant, E106Q. Can you tell the difference? That's the point. Ideally, the mutant should be identical to the wild-type protein except for that one amino acid difference. The mutant needs to fold in exactly the same way. Otherwise, it isn't clear how the activity of the two proteins could be compared, because the incorrectly folded one won't be viable as an enzyme. Consequently, researchers may compare the structures painstakingly, looking at each dihedral angle along the backbone to ensure that the two proteins adopt highly similar conformations.
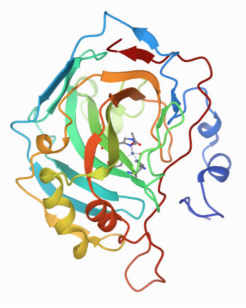
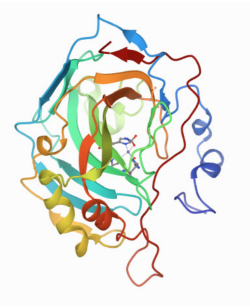
Quantifying the change in reactivity after a mutation is pretty straightforward using enzyme kinetics. We might observe a drop in kcat, or an increase in Km, or some combination of the two, resulting in a drop in efficiency of the enzyme.
Problem DI5.4.
Indicate whether the mutation resulted in a change in substrate binding, a change in the catalytic step, or whether the targeted amino acid played a minor role in catalysis.
a) WT: kcat = 4.28 x103 s-1; Km = 0. 0039 K106L: kcat = 1.1 x102 s-1; Km = 0.0043
b) WT: kcat = 6.8 x103 s-1; Km = 0.015 K106L: kcat = 7.1 x103 s-1; Km = 0.016
c) WT: kcat = 3.5 x104 s-1; Km = 0.0027 K106L: kcat = 3.2 x104 s-1; Km = 0.185
Crystal structures: RCSB Protein Data Bank
PDB ID: 1F0Q
Battistutta, R., Sarno, S., De Moliner, E., Papinutto, E., Zanotti, G.,
Pinna, L.A. Crystal structure of the alpha subunit of protein kinase CK2 in
complex with the nucleotide competitive inhibitor emodin. J.
Biol. Chem. 2000, 275, 29618-29622.
PDB ID: 1CAY
Hakansson, K., Briand, C., Zaitsev, V., Xue, Y., Liljas, A. Wild-type
and E106Q mutant carbonic anhydrase complexed with acetate. Acta
Crystallogr. D Biol. Crystallogr. 1994, 50,
101-104.
PDB ID: 1CAZ
Hakansson, K., Briand, C., Zaitsev, V., Xue, Y., Liljas, A. Wild-type
and E106Q mutant carbonic anhydrase complexed with acetate. Acta
Crystallogr. D Biol. Crystallogr. 1994, 50,
101-104.
This site is written and maintained by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (with contributions from other authors as noted). It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
Send corrections to cschaller@csbsju.edu
Navigation:
Back to Determination of Mechanism
Back to Structure & Reactivity