Structure & Reactivity in Chemistry
Structure-Property Relationships
SP13. Intermolecular Attractions and Protein Structure
Intermolecular attractions contribute a crucial part to the behaviour of everyday materials around us. In biology, attractions involving biomolecules are essential to their function. To get an idea of the role played by intermolecular attractions, such as hydrogen bonding, in biology, let's look at a common typr of biomolecule: proteins.
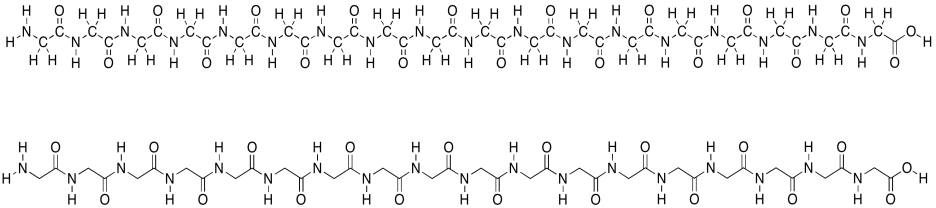
Remember that proteins are polymers made up of lots of amino acids joined together ny peptide bonds. An explicit, atom-labeled structure is shown below. In addition, there is an abbreviated, skeletal structure below that. This line drawing does not label the carbon atoms explicitly; it doesn't even show where hydrogen atoms are if they are connected to carbons. You are just supposed to know that they are there. The trade-off is a much cleaner, simpler picture.

Figure SP13.1. A simple oligopeptide shown with atom labels and in skeletal form.
In the line structure, we can easily see the zig-zag backbone of the protein chain. Periodically along that chain we can see an amide bond (N-C=O). The amide bonds have been formed by joining the amino end of one amino acid to the carboxylic end of another.
This amino acid chain isn't very long; we probably wouldn't even call it a protein, because proteins are so much bigger than that. We would just call it a polypeptide or an oligopeptide -- the latter term means there are a bunch of amino acids in this chain, but not even enough to consider it a true polymer. The particular oligopeptide we are looking at is a poly(glycine) molecule. All of the amino acids are the same monomer: glycine.
Oligopeptides, although not really proteins, are important in their own way. They are often used as signaling molecules because their small size allows them to travel relatively easily throughout an organism. Most proteins and oligopeptides are composed of different amino acids. Just to illustrate this idea, an example is shown below; it is a seven amino acid chain.
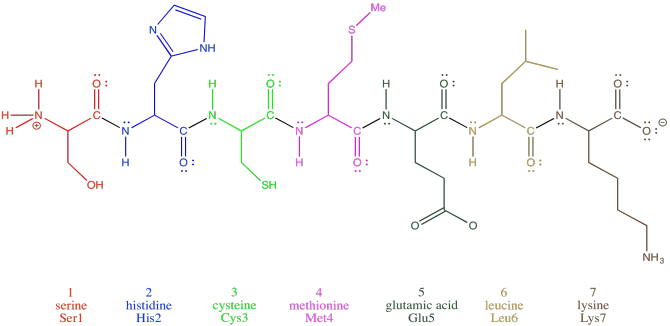
Figure SP13.2. An oligopeptide consisting of seven amino acids.
In order to help describe their structure, proteins and peptides are usually numbered, starting from the amino end of the chain (notice "oligopeptide" and "polypeptide" are a bit of a mouthful, so they often get shortened to the more general term "peptide"). Often, if a researcher wants to discuss a particular amino acid along the chain, they will list both its number and its amino acid identity. The amino acids differ from each other by their "side chain" or "residue"; that's the part hanging from the main backbone.If the water molecules don't move away from each other. For example, serine's side chain is a CH2OH group; methionine's is a CH2CH2SCH3 group. So, when you're reading the latest article in that issue of Biochemistry you've been waiting for all week, and they mention that Ser237 plays a crucial role in catalysing the formation of an important neurotransmitter, they are talking about a particular serine residue attached at the 237th position of the protein, counting from the nitrogen end.
Those protein structures can get a little bit complicated, simply because the molecules are so large. However, the basic idea of the structure is prettty simple. The same zig zag backbone runs throughout the entire molecule, and the residues are evenly spaced along the backbone. Here, we can represent those different residues as coloured dots. The structure starts to seem a little like a series of different balloons tied to a clothesline.
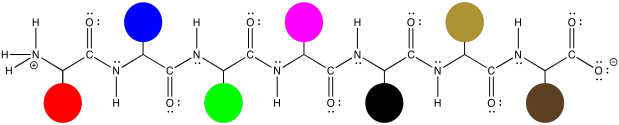
Figure SP13.3. A simplified picture of an oligopeptide shown with symbolic side chains.
That isn't to see that the structures of those side chains are not important. They are very important, and we will get back to them a little later. Sometimes, though, you have to decide what you need to focus on right now, and it might be that we want to look at the backbone without the distraction of all of those side chain structures.
In fact, even the backbone might be too much detail. Maybe we just want to keep in mind that we are dealing with a polymer, a long chain of amino acids bound together. We might fall back to an even simpler drawing of coloured beads on a string to signify that chain.

Figure SP13.4. A simple oligopeptide shown in cartoon form as a chain of beads.
Now let's just stick with the simple beads on a string picture for a minute to think about protein structure. That simple order of amino acids -- which one is at the amino end of the chain, who comes after that, and who comes after that, all the way to the carboxylate end of the chain -- is called the primary structure of the protein. The primary structure is just what amino acids are found in the protein, and in what order.
The secondary structure is a second level of detail about the protein. It begins to describe the three-dimensional shape of the structure. That's where intermolecular attractions come in to the picture. A protein, being a very long chain, can usually reach back and interact with itself. Really, we should call these intramolecular attractions, since they are within the same molecule. The interactions could involve different side chains of the protein, but frequently they also involve the backbone. Remember, the backbone has lots of carbonyl oxygen capable of donating a lone pair, and lots of amide N-H bonds capable of donating a proton, so hydrogen bonding along the backbone is very important. The cartoon below tries to suggest a chain that has looped back alongside itself, and is held in place by some sort of interactions (the dashed lines).
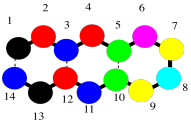
Figure SP13.5. Hydrogen bonds holding a simple oligopeptide in a fold, a feature of β-sheets.
That sort of motif, in which a protein can turn and run parallel to itself, and can be held in place by an interaction (specifically hydrogen bonds), is found in beta-sheets. Beta-sheets are very common in proteins.
Unfortunately, the cartoon doesn't really convey any details about the intramolecular attractions that hold it in place. To see that, we should take a look at a more detailed example. Here we have that polyglycine oligomer again, and we are using it to illustrate hydrogen bonding between two chains to form a beta-sheet. In this case, we have drawn the two peptides as two separate molecules. However, as in the cartoon, we could also have one long peptide that turns back to run alongside itself.
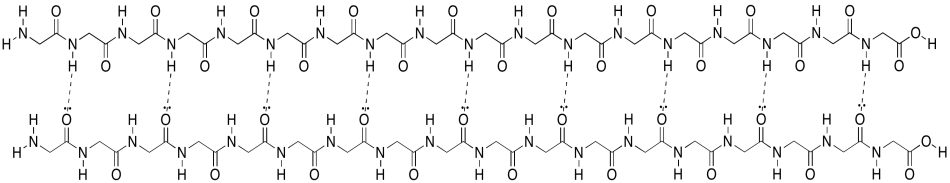
Figure SP13.6. Interactions between neighbouring sections of a peptide chain forming a parallel β-sheet.
Note that, in the example above, the two chains are running in the same direction (the amino ends are on the left and the carboxylate ends are on the right). That's an example of what's called a parallel beta-sheet. You can also have an anti-parallel beta sheet, in which the two chains are running in opposite directions. That is illustrated in the following diagram, using our simplified balloons-on-a-clothesline drawing.
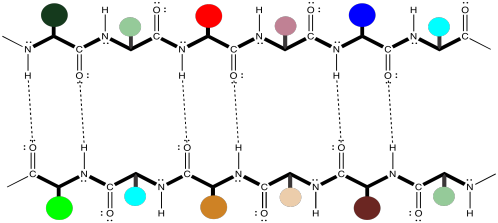
Figure SP13.7. A section of anti-parallel β-sheet shown with cartoon side chains.
The hydrogen bonds are spaced slightly differently in these two cases. Once again, these might be two different peptide chains interacting with each other, but it is very common for one chain to undergo a turn and reach back to hydrogen bond with itself. That would put the two sections of chain running antiparallel to each other. This idea is illustrated in the simple cartoon below.
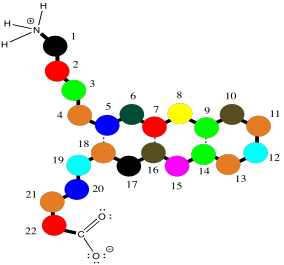
Figure SP13.8. An anti-parallel β-sheet in a short oligopeptide.
The earlier case, of a parallel beta-sheet, is also possible in a single chain; it simply involves a much longer turn, so that the chain loops around to run in the same direction as the original section. This case is illustrated in the cartoon below.
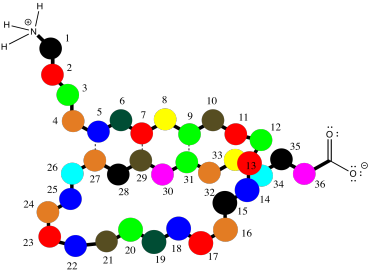
Figure SP13.9. A parallel β-sheet in a slightly longer oligopeptide wrapping back on itself.
Beta sheets, of both kinds, are very common in proteins. So are alpha-helices, in which a peptide chain coils around like a spiral staircase. It is held in that shape, once again, by hydrogen bonding along the backbone of the protein.
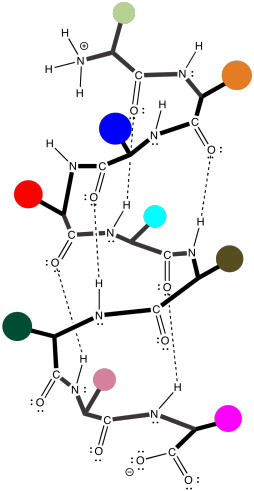
Figure SP13.10. An α-helix in a short oligopeptide.
Beta-sheets and alpha-helices are two common examples of secondary structure. They are units of structural organization that frequently show up in protein structures. Look at the following example; it's a picture of a real crystal structure result for a protein. Researchers took this protein (in this case, it's called a GTP Binding Protein) and carefully got it into a crystalline form. Then they shone X-rays through the crystal. By analysing how the X-rays were scattered as they passed through the crystal, they could go back and determine where all of the atoms were. For that, they could get this "space-filling" picture, showing the positions of all the atoms.
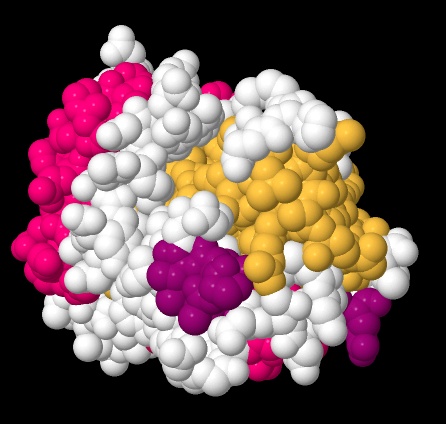
Figure SP13.11. A space-filling model of GTP-binding protein, based on X-ray data.
That picture is a little complicated, but each round ball in that mass represents a carbon, oxygen, nitrogen, or sulfur (hydrogen atoms are usually too small to spot using X-rays). The colours were added by the researchers to keep track of parts of the structure they were interested in, and, you'll see in a minute, the secondary structures.
If we go to a simpler model, we will ignore individual atoms and just trace out that backbone chain of the protein. Things will suddenly look much less complicated.
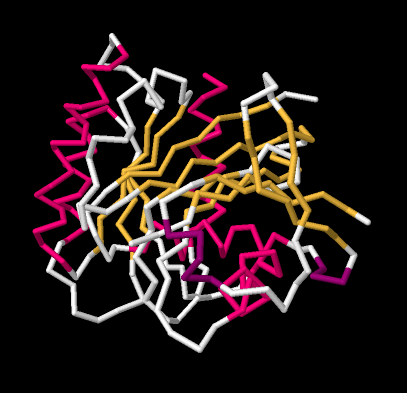
Figure SP13.12. A wireframe model of GTP-binding protein, based on X-ray data.
Now, if you take a look at the pink part on the left side, you can almost see how the backbone forms a coil, like a spiral staircase. That's an alpha helix. And those yellow zig-zags in the middle appear to be tracking each other. As one chain zigs up, so does its neighbour. When one chain zigs down, its neighbour follows. That's because they are connected. They are forming a beta sheet.
So the researchers simplify further, replacing part of the backbone with a pink helix. They replace other parts of the backbone with yellow ribbons; where the yellow ribbons run alongside each other, they are forming beta sheets. The arrows at the ends of the ribbons point towards the carboxylate end of the chain.
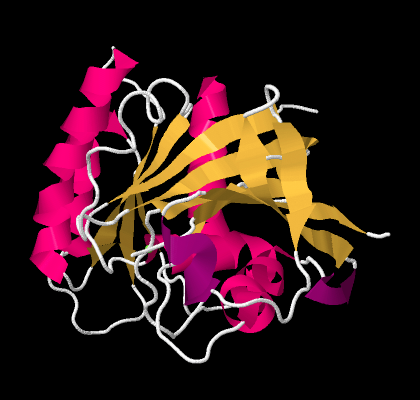
Figure SP13.13. A ribbon model of GTP-binding protein, based on X-ray data.
So a protein can contain both alpha helices and beta sheets. It can also contain less organized loops and turns. That higher level of structure -- how are the beta sheets and alpha helices and other things organized in space -- is called the tertiary structure. In a very simple cartoon, the protein might start with a length of chain, go into a beta sheet, run through another open length of chain or a loop, and finish with an alpha helix. The overall shape it forms is the tertiary structure.
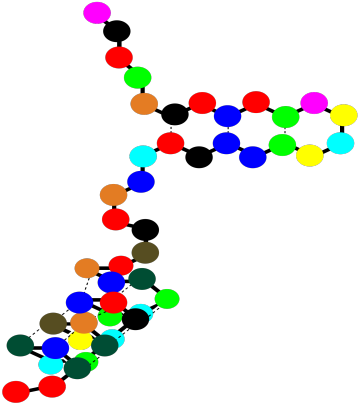
Figure SP13.14. A string-of-beads cartoon of a small protein showing tertiary structure.
There is one more level of structure in a protein, although not all proteins have this feature. It is called the quaternary structure. A quaternary structure is really a cluster of more than one protein, stuck together. Below, the cartoon shows a dimer. Two identical tertiary structures are held together by intermolecular attractions. QUaternary structure is an example of what's called a "supramolecular assembly". That's when two or more molecules come together and hold on tightly, forming a bigger structure that's bigger than a molecule.
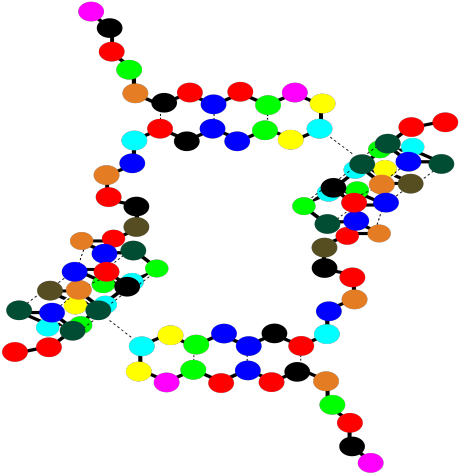
Figure SP13.15. A string-of-beads cartoon of two small protein sub-units showing quaternary structure.
Notice that there are gaps within that cartoon structure. Those gaps can be very important. Usually a protein packs very tightly and doesn't leave many spaces; when a space is there, it is often for a specific reason. A gap in a protein can hold another molecule. The other molecule might be another protein that arrives and "docks" temporarily while some important task is performed. It might also be a small molecules that needs to bind with the protein to accomplish some goal.
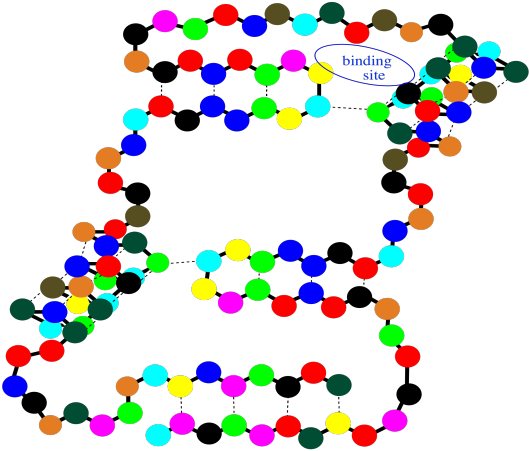
Figure SP13.16. A string-of-beads cartoon of a small protein showing an empty binding site.
These binding sites are crucial for your survival as an organism. At the same time, they are also exploited by physicians when they prescribe a medication. The drug that the doctor gives you, more often than not, binds in a specific place in a specific protein, tweaking its activity a little. Usually it gets the protein to slow down and take a break, but in some cases it may pep the protein up a little bit and make it work harder. Of course, whatever the small molecule it, and whatever it's doing there, it binds into that "active site" through intermolecular attractions.
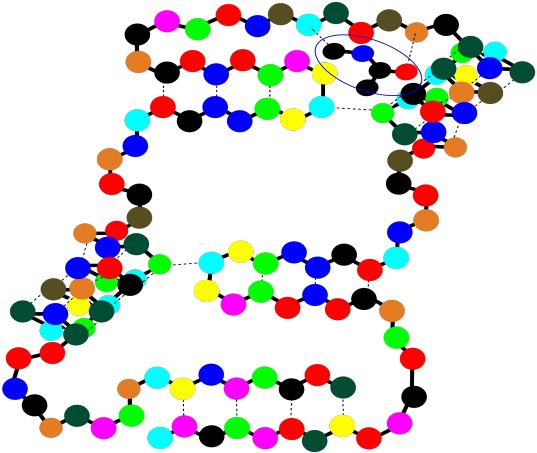
Figure SP13.17. A string-of-beads cartoon of a small molecule docking in a binding site on a protein.
That's it for protein structures. Let's just take a minute, though, to get to know the amino acids that make up proteins. The amino acids are usually divided into four different categories, partly based on their potential for intermolecular attraction. Here are the nonpolar amino acids. They are sometimes called the hydrophobic amino acids, because they can't hydrogen bond with things like water. They are shown here connected into a peptide chain, so that you can see what they would look like in a protein.

Figure SP13.18. The hydrophobic amino acids.
In contrast, the following group shows the members of the polar amino acids. They are much better at hydrogen bonding and other polar interaction, such as ion-dipole interactions.
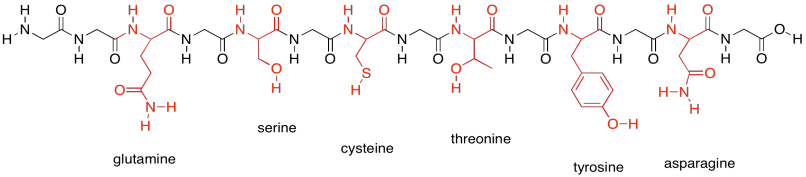
Figure SP13.19. The neutral polar amino acids.
There are a few polar amino acids that were not included in that last group because they are special. These are the basic amino acids. There are just three of them.
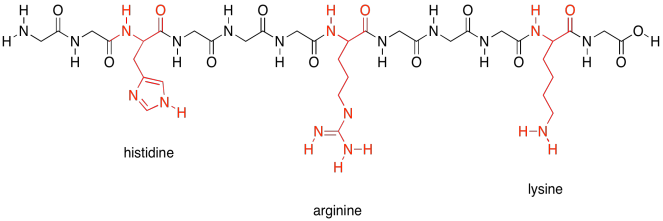
Figure SP13.20. The basic amino acids.
Finally, there are two more special polar ones. These are the acidic amino acids.
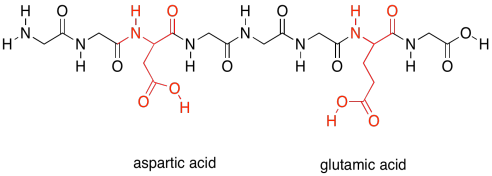
Figure SP13.21. The acidic amino acids.
Those last two groups are special because each one actually has two possible structures. They will vary depending on their environment. The basic amino acids are each able to pick up an extra proton (an H+ ion). That's very important; they help the protein carry out lots of important jobs by moving protons around.
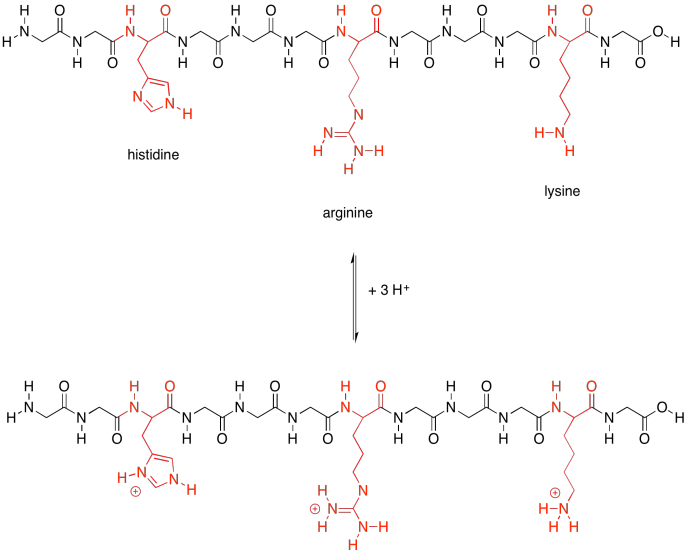
Figure SP13.22. The basic amino acids and their conjugate acids.
The acidic amino acids, on the other hand, can each give away a proton. They are also needed to carry out different tasks by the protein.
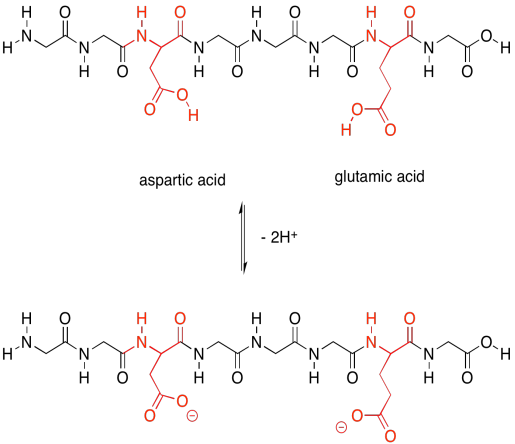
Figure SP13.23. The acidic amino acids and their conjugate bases.
Not only that, but the acidic amino acids usually have a negative charge on their side chains, because usually they are in the state in which they have lost a proton. The basic amino acids often have positive charge on their side chains, for similar reasons: they are usually in the state with an extra proton. That means that these two sets of amino acids are capable of additional intermolecular attractions, both within the protein structure and with other molecules that may come along and bind to the protein.
Problem SP13.1.
Intermolecular attractions play a crucial role in other biomolecules, such as DNA. A DNA "molecule" is really a supramolecular assembly of two separate molecules. Show how the following DNA base pairs would hold onto each other.
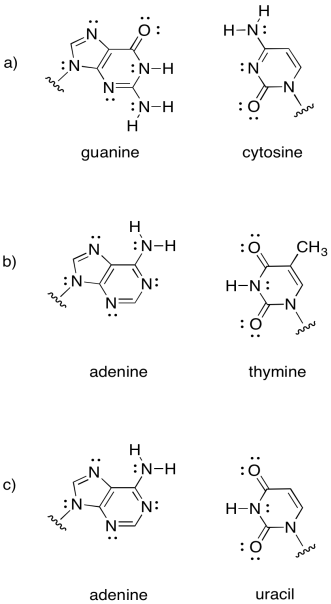
Problem SP13.2.
Intermolecular attractions can be important in other biomolecules, such as cellulose. Show how the following cellulose chains could hold onto each other.
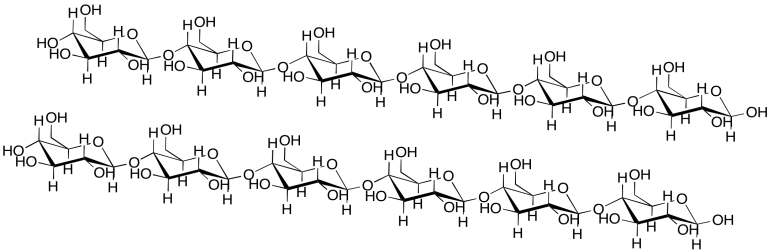
Problem SP13.3.
Cellulose is an industrially important commodity: we use it to make clothing and money (usually from cotton) as well as paper (usually from wood), for example. The pulp & paper industry expends a great deal of energy processing wood in order to make paper, with a major goal being the removal of lignin (which makes up about 15-30% of most woods, compared to about 30-50% cellulose content). The following drawing shows two chains of cellulose and a chain of lignin. Why would paper companies want to remove the lignin?
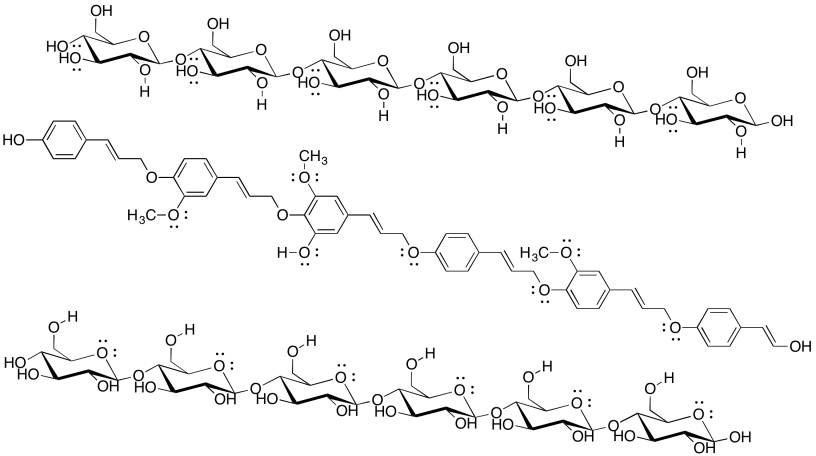
Problem SP13.4.
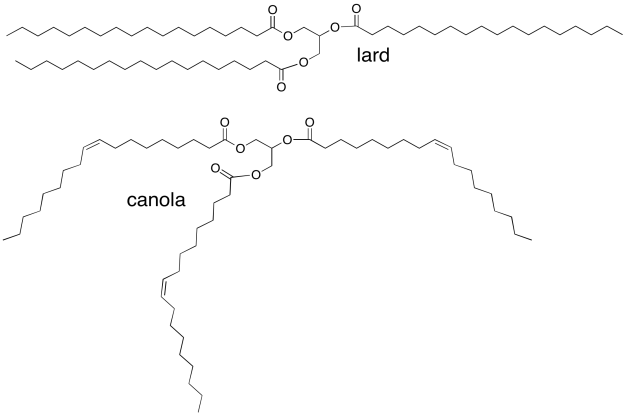
Vegetable oils, such as canola oil, are high in "unsaturated" fatty acids, in which the fatty acid chains in the triglyderides contain C=C double bonds. For example, canola oil contains about 60% oleic acid, an 18-carbon chain with a double bond in the middle. Animal fats, such as lard, contain mostly saturated fatty acids (with no C=C double bonds). Lard contains about 30% stearic acid; it's the same chain length as oleic acid but without the double bond. Why is canola an oil, whereas lard is a solid?
Problem SP13.5.
Identify the following pictures as illustrations of either alpha helices or beta sheets.
a) 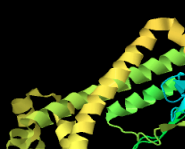 b)
b) 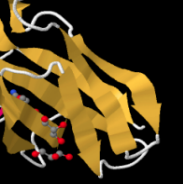 c)
c)  d)
d) 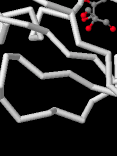 e)
e) 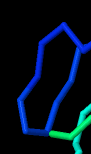
f) 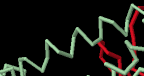 g)
g)  h)
h) 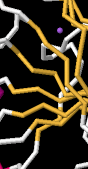 i)
i)  j)
j) 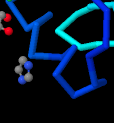
X-ray crystal structures for space-filling, backbone, and cartoon illustrations: Filchtinski, D.; Sharabi, O.; Ruppel, A.; Vetter, I.R.; Herrmann, C.; Shifman, J.M. What makes Ras an efficient molecular switch: a computational, biophysical, and structural study of Ras-GDP interactions with mutants of Raf. m>J. Mol. Biol. 2010 399: 422-435. Images obtained via RCSB Protein Data Bank (3KUD).
This site was written by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with other authors as noted on individual pages. It is freely available for educational use.
 Structure & Reactivity in Organic, Biological and Inorganic Chemistry
by Chris Schaller is licensed under a
Creative Commons Attribution-NonCommercial 3.0 Unported License.
Structure & Reactivity in Organic, Biological and Inorganic Chemistry
by Chris Schaller is licensed under a
Creative Commons Attribution-NonCommercial 3.0 Unported License.
Send corrections to cschaller@csbsju.edu
Navigation: