Reactivity in Chemistry
Nitrogen Reduction
NF3. Nitrogenase
A number of organisms take part in the nitrogen cycle, but from the point of view of chemistry, the diazotrophs have received the most attention. That's partly because their conversion of atmospheric nitrogen to ammonia mirrors the twentieth century marvel of the Haber-Bosch Process, which has been responsible for a tremendous increase in global food production over the last century.
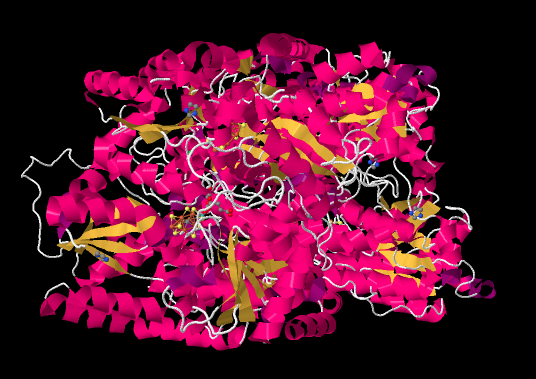
The ammonia-manufacturing center of the diazotroph is nitrogenase. The picture below is taken from an X-ray crystal structure of the enzyme (reference cited at the bottom of the page). Rather than showing the individual atoms, as X-ray structures normally do (except hydrogens, which are too small to detect), the structure is depicted in cartoon form. The cartoon image of a protein is meant to convey key substructures: the pink parts are α-helices, the yellow ribbons are β-sheets, and the white threads are loops.

Figure NF3.1. The experimentally observed structure of nitrogenase (Spatzal et al).
If we delete all of the protein from the image -- that is, the chain of amino acids that make up nitrigenase -- then we are left with a few "ligands". Ligands in this context are simply other molecules or ions found within the protein; often the ligands are bound to amino acid side chains. Looking at the ligands below, we can see that nitrogenase is actually a dimer. The group of molecules in the left half of the picture are the same as the group of molecules in the right half of the picture, although they are rotated upside down with respect to the others.
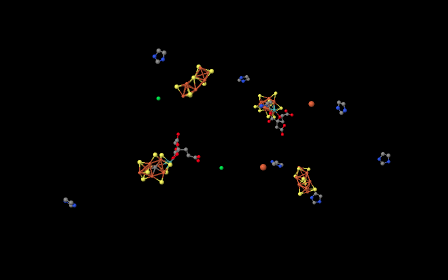
Figure NF3.2. Some important electron carriers within nitrogenase (Spatzal et al).
A couple of the structures in the picture above are metal ions; they look like little balls. To get a better view of the other structures, let's zoom in a little closer.
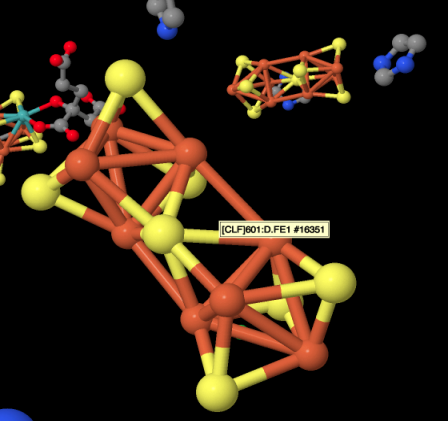
Figure NF3.3. An FeS cluster in nitrogenase (Spatzal et al).
That's an iron sulfur cluster photobombing the picture. The red spheres are colour-coded for iron, whereas the yellow sphered are colour coded for sulfur. The bars between the atoms may or may not represent bonds; X-ray diffraction doesn't really detect bonds, just the highly electron-dense atoms, but the software inserts bonds when it detects atoms that are close together. The iron-sulfur bonds are certainly real. The red bonds shown between the iron atoms probably are not real, although there is a possibility of magnetic coupling between the iron atoms within the cluster. A drawing of the cluster is provided below. The drawing also includes additional sulfur donors on the iron atoms, but those are from cysteine residues in the protein, which has been rendered invisible in the picture above.
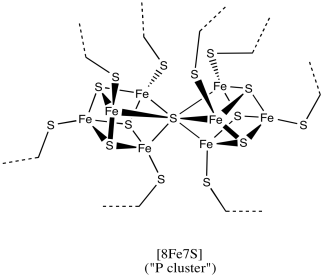
Figure NF3.4. A drawing of an FeS cluster in nitrogenase.
This particular cluster contains 8 iron atoms and 7 sulfide ions (S2-); it's referred to as an [8Fe7S] cluster for short. Iron sulfur clusters are actually pretty common in biology. Probably the most common is a [4Fe4S] cluster, although [3Fe4S] and [2Fe2S] clusters are often seen, too. Iron sulfur clusters are usually charged. The amount of charge on the overall clusters depends on the oxidation states of the iron atoms. The oxidation states can change because these clusters function as electron relays. They can accept an electron from elsewhere in the protein and send it on to where it is needed.
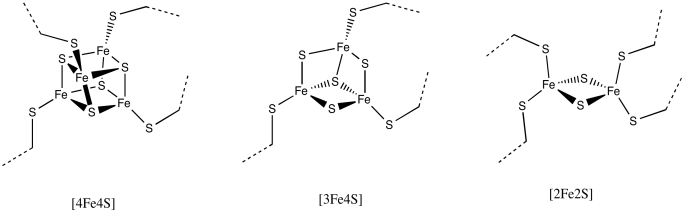
Figure NF3.5. Drawings of other, more common FeS clusters.
Problem NF3.1.
Indicate the charges on the following clusters.
a) [4Fe4S], with two Fe2+ and two Fe3+
b) [3Fe4S], with two Fe3+ and an Fe2+
c) [2Fe2S], with one Fe2+ and one Fe3+
There are also a couple of histidines visible in the picture above. They were probably introduced when the researchers were growing the crystals. A far more interesting structure is barely visible on the left. That's actually the iron-molbdenum or Fe-Mo cofactor, also called the M cluster. It's the site of nitrogen reduction in the enzyme. A much clearer view of this structure is provided below. Because we are looking at a dimer, we can see a second M cluster behind.
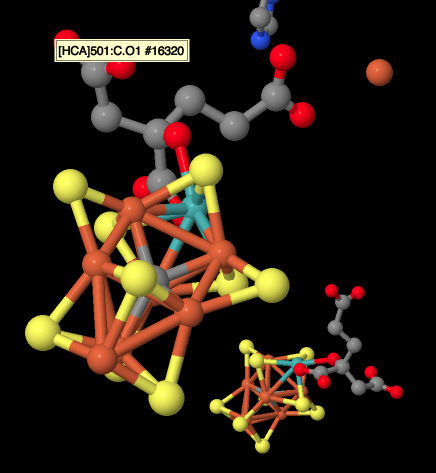
Figure NF3.6. A very special FeS cluster in nitrogenase; it also contains Mo (Spatzal et al).
A drawing of the M cluster is shown below. It bears some resemblance to [8Fe7S]. This time, the atom in the center is believed to be a carbon. A trio of additional sulfides bridge between the two cubes, and in one corner an iron atom has been replaced by a molybdenum.
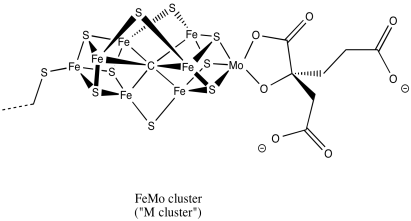
Figure NF3.7. A drawing of the Fe-Mo cofactor in nitrogenase.
Here is a picture of the M cluster from a different angle. The molybdenum with its attached ligand is in the upper left, coordinated by yellow sulfides. The gray atom in the middle is probably a carbon atom.
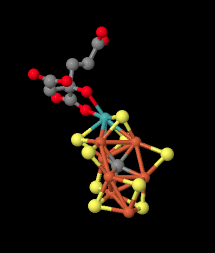
Figure NF3.8. Another image of the Fe-Mo cofactor in nitrogenase (Spatzal et al).
The mechanism by which nitrogenase reduces nitrogen to ammonia is likely somewhat different from the mechanism in the Born-Haber Process. Rather than treatment with hydrogen gas, under biological conditions the mechanism is likely to involve reduction via individual electrons and protons. Exactly how the electrons and protons arrive at their destination is an interesting question.
One possibility is that the dinitrogen is reduced one electron and one proton at a time. The FeS clusters would be able to deliver electrons via an electron relay running through the cell. Amino acid residues in the protein would be able to shuttle protons in as needed.
Problem NF3.2.
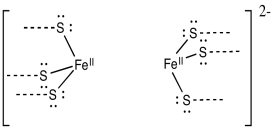
Provide a mechanism for the reduction of dinitrogen, N2, to diazene, HN=NH, via alternating additions of electrons and protons. Assume the following model is the site of the reaction, with appropriate electron and proton donors nearby.
Alternatively, the process could more closely resemble the Haber-Bosch Process, using classic "organometallic" reaction mechanism such as oxidative addition, insertion and reductive elimination. In fact, recent evidence suggests H2 may be produced by some nitrogenases under biological conditions. That obsevration is consistent with the operation of this type of mechanism.
Problem NF3.3.
Show how protonation of an iron atom is equivalent to an oxidative addition.
Problem NF3.4.
Provide a mechanism for the reduction of dinitrogen, N2, to diazene, HN=NH, via a series of organometallic steps (association/dissociation, oxidative addition/reductive elimination, insertion/elimination).

X-Ray structure: Spatzal, T., Perez, K.A., Einsle, O., Howard, J.B., Rees, D.C. Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase. Science 2014, 345, 1620-1623. Images obtained via the RCSB Protein Database (4TKU).
This site was written by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with contributions from other authors as noted. It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
Navigation: