Reactivity in Chemistry
Oxygen Binding & Reduction
OR3. Oxygen Reduction
Molecular oxygen is exploited in a variety of ways in biology. Apart from its central role in metabolism, it also plays a crucial part in the modification of many compounds through the incorporation of oxygen atoms into the structure.
For example, the cytochromes P450 form an important class of enzymes involved in the oxidation of substrates, chiefly in the liver. One atom of oxygen from an O2 molecule is incorporated into the substrate molecule; the remaining oxygen atom is converted to water. This type of enzyme is called a "monooxygenase" because of the addition of one oxygen atom from O2 into the substrate.
The addition of oxygen atoms to molecules, typically in the form of hydroxyl groups, is vitally important. The reaction may have evolved for a number of reasons. A key reason is to increase the water-solubility of small organic molecules, which are then more easily excreted via the urine or broken down via subsequent reactions. Hydroxylation lets us get rid of foreign substances. Otherwise, these hydrophobic molecules would build up in the tissues.
The cytochrome P450 pathway is a major avenue for the breakdown and excretion of pharmaceuticals, for instance. We can get rid of these substances after they have done their job. In other cases, pharmaceuticals are not active until they are hydroxylated; the reaction acts as an "on" switch. In still other cases, hydroxylation is a dangerous complication, converting a helpful pharmaceutical into a toxin.
Exactly how does the O2 get broken into pieces suitable for incorporation into other molecules? It's a very complicated problem. There is an entire field of chemists who study "small molecule activation", which refers to the breakdown of things like O2, N2, CO or methane for their subsequent conversion into other compounds. In fact, there is an entire field of chemists and biochemists who just focus on cytochrome P450, trying to learn more about how it works. They do so both to understand more about a biological system that is related to human health and to gain insight into how to improve industrial processes involving oxidation of substrates.
Some things are pretty well understood. We'll take a general look, leaving out some important details so things aren't too overwhelming.
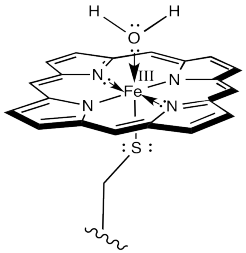
The first step is just the binding of dioxygen to a metal. In cytochrome P450, that iron atom looks very much like the one in hemoglobin. In its resting state, it is formally an Fe(III) ion in a porphyrin ring, but with an axial cysteine donor instead of a histidine. In addition, a water molecule is coordinated to form an octahedral complex.

Figure OR3.1. The active site of cytochrome P450 in its resting state.
That leaves no place for the dioxygen to bind. However, once the substrate enters the enzyme, a conformational change results in loss of the water molecule. After that, an electron is delivered from a cofactor, leading to an Fe(II) complex.
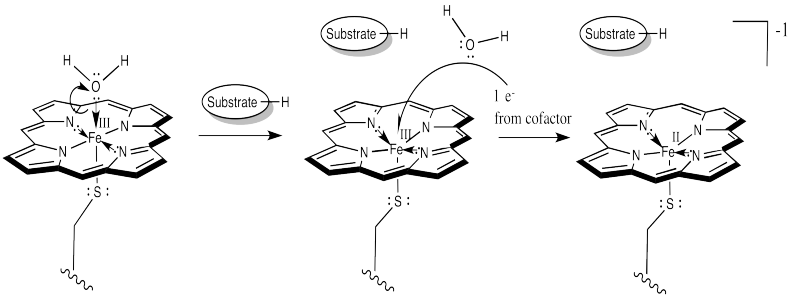
Figure OR3.2. Cytochrome P450 is reduced in its activated state.
At this point, things are looking a little more like hemoglobin. Just as in hemoglobin, the O2 binds to the iron, which immediately transfers an electron to the bound dioxygen, forming an Fe(III) superoxide complex.
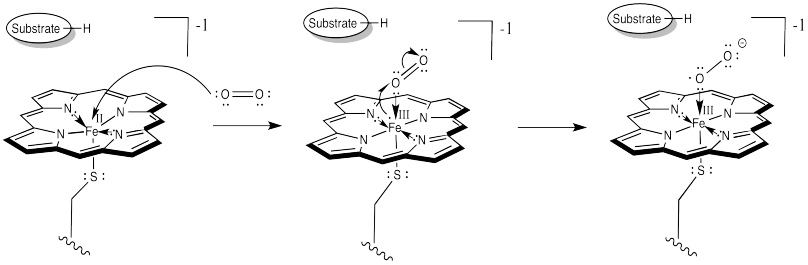
Figure OR3.3. The activated cytochrome P450 binds and reduces dioxygen.
The difference is that things do not stop there. The addition of a second electron from a cofactor tips things a little further. One more electron turns the superoxide ion into a peroxide ion.
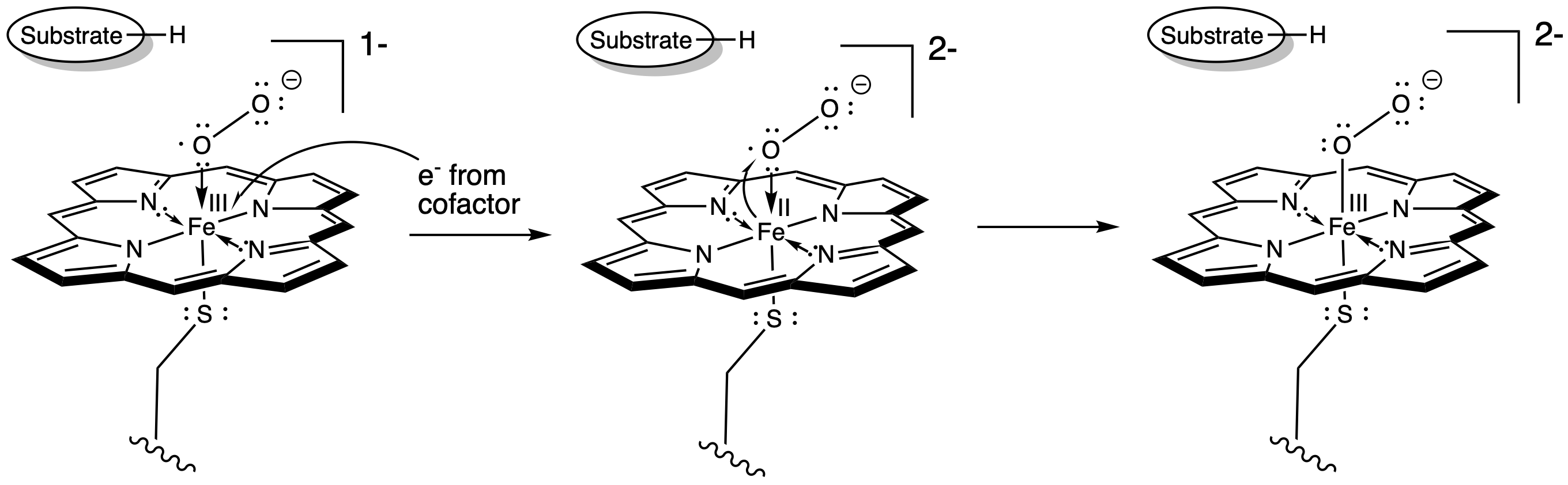
Figure OR3.4. Further electron donation to cytochrome P450 leads to further reduction of dioxygen.
At this point, we have completely severed the first of the two O=O bonds. We are almost there. The subsequent addition of two protons leads to the formation of a water molecule. Now we have one oxygen atom bound to iron.
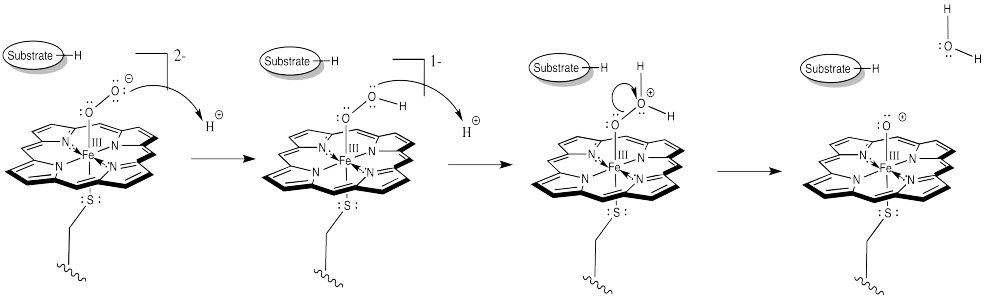
Figure OR3.5. Proton transfer to the peroxide complex and subsequent dehydration leads to an iron oxo complex.
How do we think about that species? We can think about several resonance structures. The electron-deficient oxygen is attached to an iron atom. The iron possesses a reservoir of electrons. It can donate one to the oxygen atom. It might even donate two electrons, making an iron (V) oxide complex, but that might be going too far.
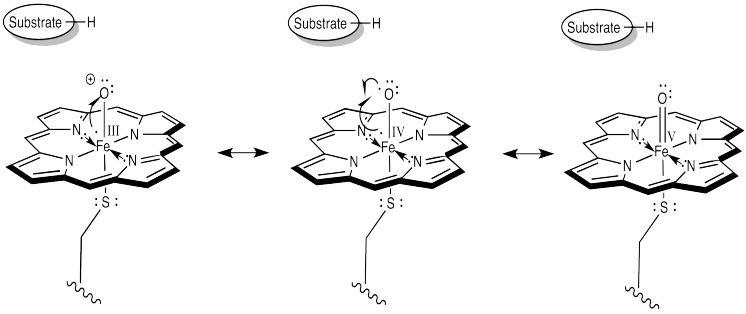
Figure OR3.6. Resonance structures of an iron oxo complex.
In the resonance structure with iron (IV), we see an oxygen atom with a single, unpaired electron. This species is called a radical. Radicals are notorious for ripping hydrogen atoms from other molecules. That just makes a new radical. This is the trouble with radicals; they are hard to get rid of, because they are always making more. It also makes an iron hydroxy complex, which can combine with the radical in a "rebound" step to form the hydroxylated compound.
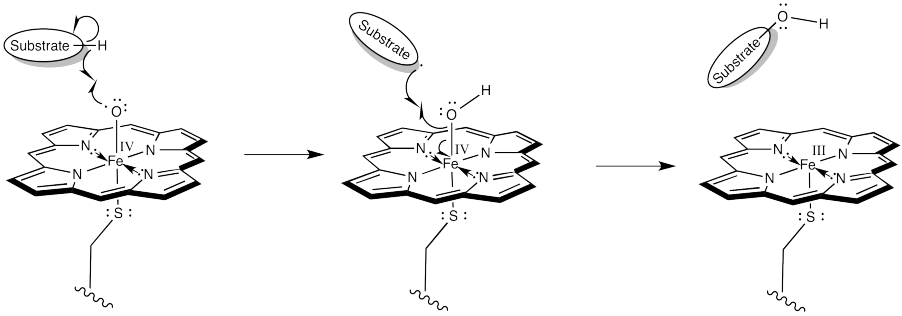
Figure OR3.7. Carbon-hydrogen bond activation and hydroxylation by an iron oxo complex.
We will see more of this kind of event in the chapter on radicals.
Problem OR3.1.
Fill in the missing intermediates in the following scheme.
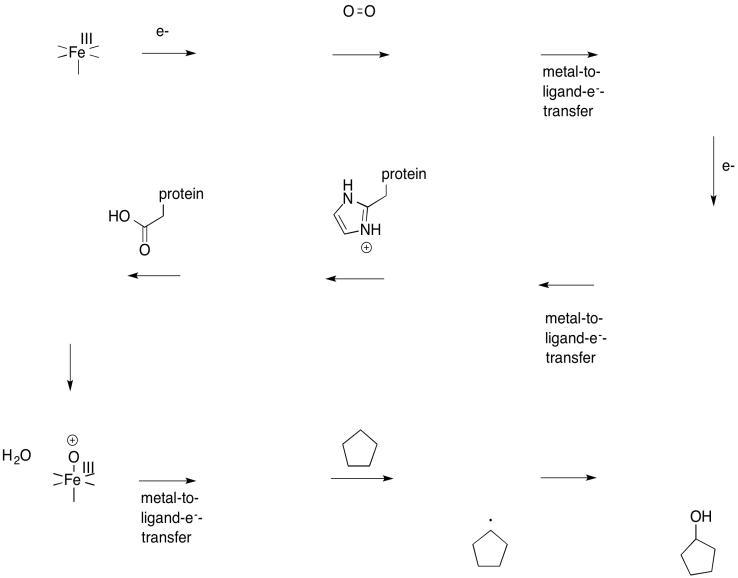
Problem OR3.2.
The Karlin lab (Johns Hopkins) has developed a series of model compounds for copper-containing monooxygenases (J. Am. Chem. Soc. 2014, 136, 8063-71).
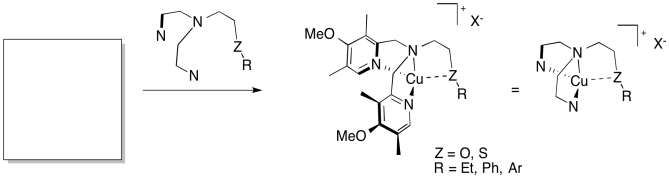
a) Fill in a reasonable starting material that you might find in the chemistry
stockroom.
b) Reaction with O2 gives dimeric copper
complexes (LnCu2) of two types: with one bridging peroxide
anion or with two bridging oxide anions. Draw both dimers.
c) O2 is reduced / oxidized (select one) by _____ (number of ) electrons to make peroxide anion.
d) O2 is reduced / oxidized (select one) by _____ (number of ) electrons to make oxide anion.
e) Identify the oxidation states of the copper ions in the two dimers.
f) In the ligand, if the group ZR = SEt, the reaction forms the peroxide bridge. If ZR = OEt, it forms the oxide bridge. Suggest a reason why.
g) With Z = S, the group R both influences which dimer forms and affects the Cu-S bond. Predict the combination of features found in each product and provide an explanation.
R = Et peroxide / oxide Cu-S bond / no Cu-S bond
R = Ph peroxide / oxide
Cu-S bond / no Cu-S bond
This site was written by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with contributions from other authors as noted. It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.