Reactivity in Chemistry
Photosynthesis
PS6. Harvesting the Photons: Photosystem I
Having already generated the proton gradient used for ATP generation, what is there left to do?
Remember back to that high school biology class when you first saw the equation for photosynthesis.
6 CO2 (g) + 6 H2O (l) → C6H12O6 (s) + 6 O2 (g)
We can break that into two half reactions. The first is the oxidation of water.
2 H2O (l) → O2 (g) + 4 H+ (aq) + 4 e- (aq)
The other half is the reduction of carbon dioxide.
6 CO2 (g) + 24 H+ (aq) + 24 e- (aq) → C6H12O6 (s) + 6 H2O (l)
Although these two half-reactions are physically separated from each other, the electrons must still get from one to the other in order to complete the full reaction. We have already accomplished the first reaction and now have electrons in hand to carry out the second. We are going to need a biological reducing agent, like NADH, to carry out the second. In this case, we will use a closely-related compound, NADPH.

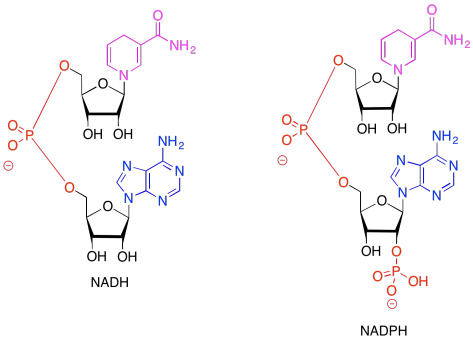
Figure PS6.1. Comparative structures of two related electron carriers: NADH and NADPH.
In both of the drawings above, the purple portion of the compound is where the electrons are accepted. The black portion is sugar, the blue is a purine base, and the red is the phosphate part.
Problem PS6.1.
Because NADH and NADPH are electron carriers, it is important to recognise both reduced and oxidised forms. Given the reduced forms of NADH and NADPH shown above, draw the oxidised forms, and show where the :H- is incorporated in the molecule in the reduced form.
The problem is, NADPH is a relatively high-energy electron carrier -- remember, NADH was at the uphill end of the electron transport chain in oxidative phosphorylation -- and our electrons have just ridden their sled to the bottom of the hill. If we are going to make NADPH, we will need to tug the electrons back uphill again. That's essentially the job of photosystem I, shown below.
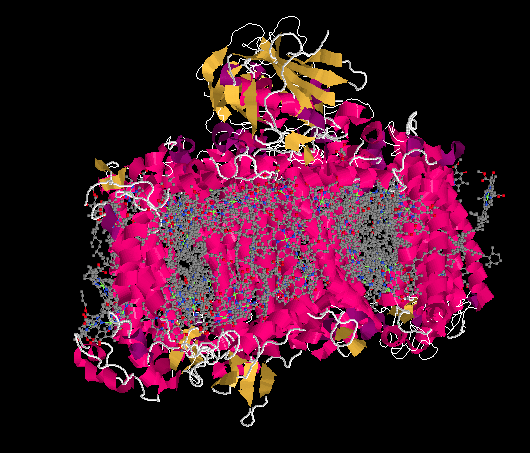
Figure PS6.2. Results from an X-ray crystal structure of photosystem I in the marine cyanobacterium Synechococcus elongatus.1
The ligand-only view of the crystal structure reveals a picture that should be somewhat familar now: a host of antennae molecules forming a light-harvesting complex. Once again, the antennae molecules include mostly chlorophylls, as well as carotenoids such as carotene and lycopene, plus a few xanthins like violaxanthin; the exact molecules vary depending on whether we are looking at plants, algae, or bacteria.
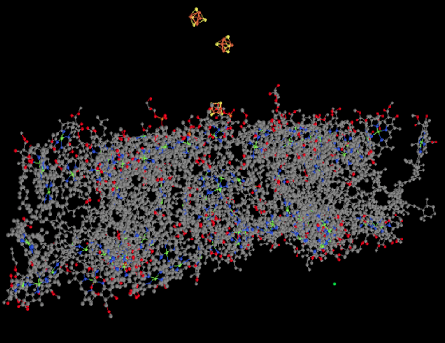
Figure PS6.3. Ligand-only view of an X-ray crystal structure of photosystem I.
Also visible in the ligand-only view is a group of FeS clusters. These clusters play a role in the electron-transport chain of photosystem I, conducting the electron toward the stroma, where they will be used to reduce NADP+ to NADPH. That biological reducing agent, closely related to NADH, will be used to help reduce carbon dioxide.
Let's pause for a moment. Most of the X-ray crystal structures we have seen have come from bacteria. Of course, we are much more familiar with the idea of photosynthesis in plants. Are the proteins and processes involved in photosynthesis in one anything like the other? For comparison, here is photosystem I from a spinach plant.
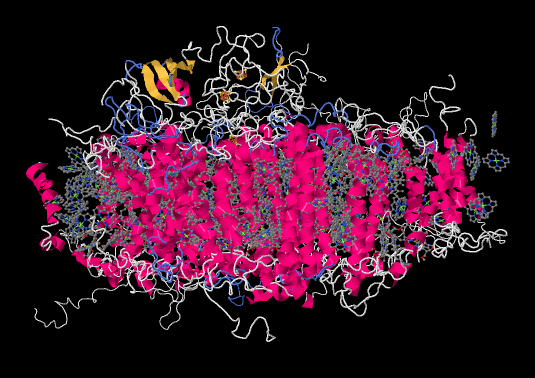
Figure PS6.4. Results from an X-ray crystal structure of photosystem I in spinach.2
It is roughly similar, although not identical, to the one from the bacterium. There is a broad swath of alpha helices through the middle, topped with a bundle of beta sheets. If anything, the protein from the spinach looks superficially a little simpler than the one from the bacterium. The ligand-only view, below, is strikingly similar, showing a spray of chromophores and a series of iron-sulfur clusters.

Figure PS6.5. Ligands-only view from an X-ray crystal structure of photosystem I in spinach.
Let's pick up the story where we left off when we were discussing cytochrome b6f. A single electron was being transported by the soluble, mobile electron carrier, plastocyanin. That single electron is delivered directly to the reaction centre of photosystem I. That reaction centre is quite similar to the one in photosystem II, although it absorbs photons of a slightly longer wavelength, 700 nm. The P700 centre contains an excitonic dimer, a closely-spaced pair of chlorophyll molecules that absorb the photon and undergo electron / hole separation on the two different molecules.
Just as in P680, the P700* excited state can accept an electron onto the chlorphyll partner that has the hole, PD2+*. That's where the plastocyanin delivers its electron.
Meanwhile, the part of the dimer with the extra electron, PD1-*, passes the electron on to another station on the electron transport chain. Part of the goal here is to get the electrons back to the stroma where the NADP+ is waiting. If you think about it, the electrons started out in the lumen, on a water molecule. They traveled across the thylakoid membrane in photosystem II until they joined a plastoquinone and picked up a couple of protons; those protons came from the stroma. The plastoquinones travelled back across the membrane, a stealth migration of polar H+ in the form on nonpolar PQH2, and delivered their protons to the lumen. The side-kick electrons skipped along the edge of the lumen in a plastocyanin carrier, and are now traveling back toward the stroma to that waiting NADP+. The NADPH will be produced in the stroma because that's where it will be needed for CO2 capture.
Again, just as in P680, the first electron acceptor after P700 is a single chlorophyll. This chlorophyll anion that forms is slightly downhill in energy from the one in the dimer, PD1-*. From there, the electron passes to a phylloquinone, which is just another variation on the familar quinone family.
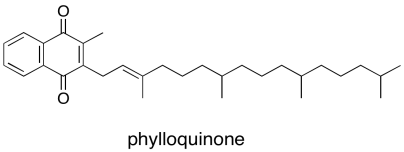
Figure PS6.6. The structure of phylloquinone.
The phylloquinone sends the electron into that series of three FeS clusters visible in the crystal structure. From there, the electron leaves photosystem I and is deposited on a ferredoxin, a small protein that binds another iron sulfur cluster. The ferredoxin is located in the stroma, where it can bind with an enzyme called ferredoxin NADP+ reductase. Like most enzymes, the name says exactly what it does: it catalyses the reaction in which ferredoxin reduces NADP+, resulting in NADPH.
The crystal structure of a ferredoxin is shown below. This particular one is not from photosystem I (it is found in a nitrogen-fixing bacterium), but it gives you the idea. The protein is simple enough that you can see that it is a dimer; the top half is the same as the bottom half, flipped over. You can also clearly see a 2Fe2S cluster at the edge of each subunit. The location of that FeS cluster near the edge of the protein is probably important in facilitating electron transfer with another protein.
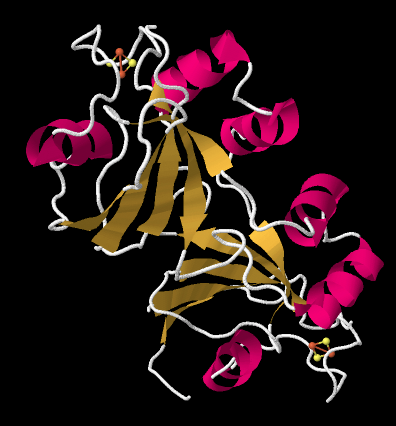
Figure PS6.7. X-ray crystal structure of a ferredoxin, a small protein containing a FeS cluster, from Azotobacter vinelandii.3
The ferredoxin is an electron carrier protein. Its job is to deliver electrons to the ferredoxin NADP+ reductase in the stroma, which will then use the electrons to make NADPH. NADPH is a biological source of hydride ion (formally, H-, or a proton plus two electrons) that in this context will be used to help reduce carbon dioxide into carbohydrate. Below, we can see an example. This is another dimeric protein with rotational symmetry: the left side is the same as the right, but turned over.
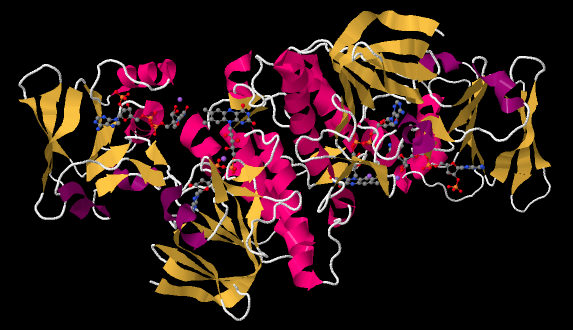
Figure PS6.8. X-ray crystal structure of a ferredoxin NADP+ reductase from Bacillus subtilis.4
If we look inside, we can see the NADP+ molecules waiting at either end of the structure, at both the far left and the far right. In addition, there is a pair of molecules that look vaguely related to NADP+ toward the middle of the picture. Those are two FAD molecules. The FAD molecules are there to conduct electrons from the ferredoxin t the NADP+.
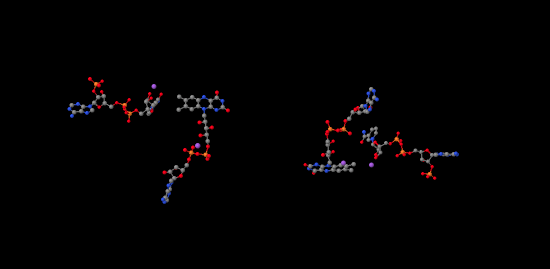
Figure PS6.9. Ligand-only view of the X-ray crystal structure of a ferredoxin NADP+ reductase.
In the picture below, the purple part of the molecule is the redox-active region. It is able to accept electrons. The black part is sugar, the blue is a purine base, and the red is phosphate.
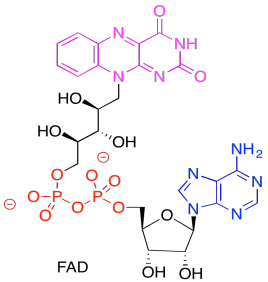
Figure PS6.10. The structure of the electron carrier, FAD.
Problem PS6.2.
Show the reduced form of FAD. Remember, the reduced form is called FADH2.
Problem PS6.3.
What role do you think the FAD plays in ferredoxin NADP+ reductase? Why doesn't the ferredoxin reduce the NADP+ directly?
Problem PS6.4.
Because photosystem II and photosystem I are part of the same electron transport chain, it is important that the reaction centre in one is getting excited just as often as the reaction centre in the other. Regulation is partly accomplished via covalent modification. For example, insufficient activity at photosystem I results in activation of a kinase, which results in release of some of the antennae molecules from the light harvesting complex in photosystem II.
a) How would this situation help to remedy the lack of parity in activity between photosystem I and photosystem II?
b) Explain how the kinase would achieve this result.
c) Suppose the opposite situation occurred: insufficient activity at photosystem II. What enzyme would probably be activated to remedy the situation?
We can summarize with a sketch of photosystem I:
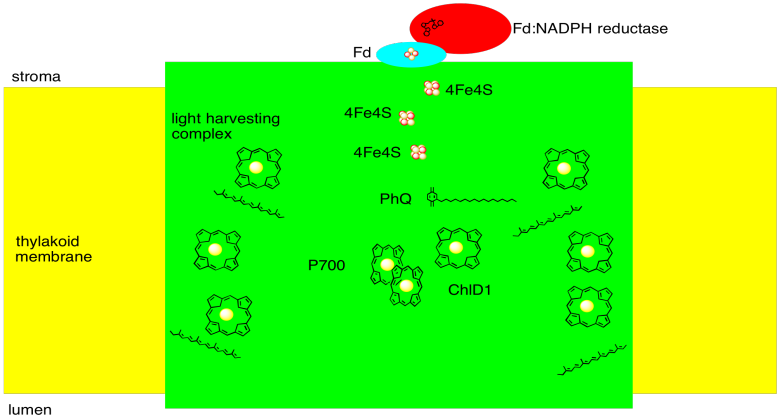
Figure PS6.6. Sketch of photosystem I.
1. X-ray crystal structures: Jordan, P.; Fromme, P.; Witt, H.T.; Klukas,
O.; Saenger, W.; Krauss, N. Three-dimensional structure of cyanobacterial
photosystem I at 2.5 A resolution. Nature 2001,
411, 909-917. Images obtained via RCSB Protein Data Bank (1JB0).
2. X-ray crystal structures: Amunts, A., Drory, O., Nelson, N. The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 2007, 447, 58-63. Images obtained via RCSB Protein Data Bank (2o01).
3. X-ray crystal structures: Schlesier, J., Rohde, M., Gerhardt, S., Einsle, O. A Conformational Switch Triggers Nitrogenase Protection from Oxygen Damage by Shethna Protein II (FeSII). J. Am. Chem. Soc. 2016, 138, 239-247. Images obtained via RCSB Protein Data Bank (5ffi).
4. X-ray crystal structures: Komori, H., Seo, D., Sakurai, T., Higuchi,
Y. Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+)
oxidoreductase and the structural basis for its substrate selectivity.
Protein Sci. 2010, 19, 2279-2290. Images obtained
via RCSB Protein Data Bank (3Lzw).
This site is written and maintained by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with contributions from other authors as noted. It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
This material is based upon work supported by the National Science Foundation under Grant No. 1043566.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Navigation: