|
Self-splicing group I intron with both exons |
The
fact that not all enzymes are proteins may be demonstrated by the
RNA self-splicing group I intron. This model depicts a complete
group I bacterial intron complexed with both the 5’- and 3’- exons.
During splicing, the intron will be removed, leaving only the exon
portion of RNA. Whereas splicing is typically catalyzed by a
ribonucleoprotein called a spliceosome, this comples catalyzes the
excision of its own intron without the addition of accessory factors
Wire Frame
Backbone
Cartoon
Molecule Characteristics
The intron uses unique RNA motifs in order to select splice sites.
An OH on the 5’-exon acts as a nucleophile, attacking a phosphate on the 3’-exon.
Highlight nucleophilic OH
Highlight nucleophilic OH and complexing heteroatoms
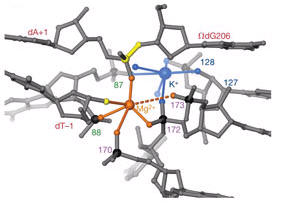
Six phosphates from three RNA strands coordinate two metal ions (probably magnesium 2+) positioned at the reactive phosphate (3’-exon).
This particular structure represents the state preceding the second step of splicing, at which the 5’-exon has been cleaved, but is still base-paired to the intron. The 3’-exon is still covalently bound to the intron, but also is helically base-paired to the intron.
Highlight 3' Exon (red)
Highlight 5' Exon (blue)
Highlight Intron (white)
Highlight nucleophilic OH, complexing heteroatoms, and both exons
Wire Frame of Splicing Site
Splicing Site wih details {intron in grey, 3' exon in red, 5' exon in blue, heteroatoms highlighted}
Figure displaying coordination of the active-site metal ions, Mg2+ and K+. These ions coordinate with the nucleophile, the leaving group, and scissile phosphorous. Note the high phosphorous density in the active site. Transition metals in this complex have been proposed to activate the nucleophile, stabilize the leaving group, and neutralize negative charges on phosphorous transition states.
Highlight Potassium Heteroatoms (pink)
Highlight Magnesium Heteroatoms (green)
Highlight All Heteroatoms
|