Biochemistry Online: An Approach Based on Chemical Logic

Metabolic Pathways
CT0. The Cell: An Overview
Henry Jakubowski
French Translation √ by Kate Bondareva
You have probably studied the cell many times, either in high school, or in college biology classes. There are many websites available that review both prokaryotic (bacterial), archeal and eukaryotic cells (see links at bottom). This tutorial is designed specifically from the viewpoint of chemistry. It explores four classes of biomolecules (lipids, proteins, nucleic acids and carbohydrates) and describes in a simplified pictorial manner where they are found, made, and degraded in the cell (i.e. their history). It focuses on eukaryotic cells, which in contrast to simpler prokaryotic cells, have internal organelles surrounded by membranes that compartmentalize chemical reactions. First a general overview of the cell is presented. Subsequent chapters will concentrate on specific biomolecules.
Let's think of a cell as a chemical factory which designs, imports, synthesizes, uses, exports and degrades a variety of chemicals (in the case of cell, these include lipids, proteins, nucleic acids and carbohydrates). It also must determine or sense the amount of raw and finished chemicals it has available and respond to its own and external needs by ramping up or shutting off production. The figure of the cell below and in other linked sites based on it was made available with the kind permission of Liliana Torres. Click on the blue hyperlinks for some of the organelles for more detailed information on them.
Figure: A Eukaryotic Cell
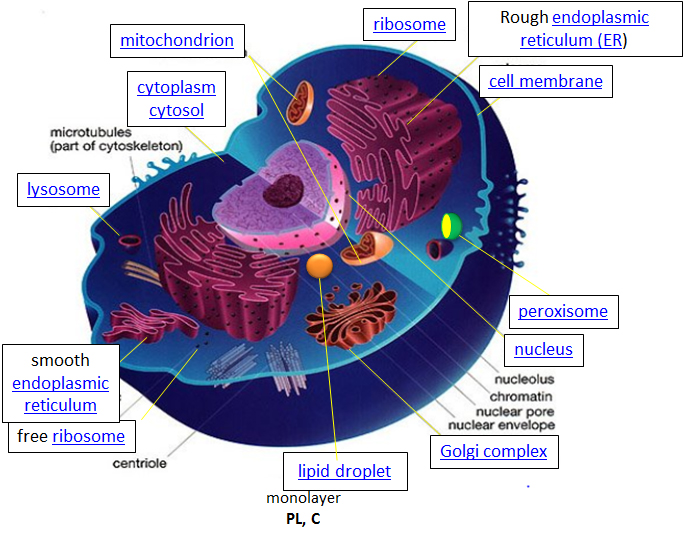
http://torresbioclan.pbworks.com/w/page/22377234/Spikefish%20About%20Cells
Use with permission from Liliana Torres . Also at http://www.animalport.com/animal-cells.html
Design - The design for a cell mostly resides in the blueprint for the cell, the genetic code, which is comprised of the DNA in the cell nucleus and a small amount in the mitochondria. Of course, the DNA blueprint must be read out (transcribed) by protein enzymes which themselves were encoded by the DNA. The genetic code has the master plan that determines the sequence of all cellular proteins, which then catalyze almost all other activities in the cell, including catalysis, motility, architectural structure, etc. In contrast to DNA, RNA and protein polymers, the length and sequence of a polysaccharide polymer is not driven by a template but rather by the enzymes that catalyzes the polymerization.
Import/Export: Many of the chemical constituents of the cell arise not from direct synthesis but from import of both small and large molecules. The imported molecules must pass through the cell membrane and in some cases through additional membranes if they need to reside inside membrane-bound organelles. Molecules can move into the cell by passive diffusion across the membrane but usually their movement is "facilitated" by a membrane receptor. Molecules can also move up a concentration gradient in a process called "active transport". Given the amphiphilic nature of the bilayer (polar head group exterior, nonpolar interior), you would expect that polar molecule like glucose would have difficulty in moving across the membrane. In fact membrane transport proteins are involved in the movement of both nonpolar and polar molecules.
- transporters, carrier proteins and permeases: These membrane proteins move specific ligand molecules across a membrane, typically down a concentration gradient. Computer simulations of the facilitated diffusion of lactose across the membrane is shown in the following link. Animation of lactose diffusion through the LacY receptor
(The link above and immediately below are from the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign. These molecular dynamic simulations were made with VMD/NAMD/BioCoRE/JMV/other software support developed by the Group with NIH support.)
- ion channels - These membrane proteins allow the flow of ions across membranes. Some are permanently open (nongated) while others are gated open or closed depending on the presence of ligands that bind the protein channel and the local environment of the protein in the membrane. Flow of ions through the channel proceeds in a thermodynamically favored direction, which depends on their concentration and voltage gradients across the membrane.
- pores: Some membranes (nuclear, mitochondria) assemble proteins (such as porins) to form large, but regulated pores. Porins are found in mitochondrial membranes while nucleoporins are found in the nuclear membrane. Small molecules can generally pass through these membrane pores while large one are selected based on their tendency to form transient intermolecular attractive forces with the pore proteins. The following link shows the diffusion of water through aquaporin. animation of water diffusion through the aquaporin channel,
- endocytosis: Very large particles (for example, LDL or Low Density Lipoproteins or viruses) can enter a cell through a process called endocytosis. Initially the LDL or virus binds to a receptor on the surface of the cell. This triggers a series of events which leads to the invagination of the cell membrane at that point. This eventually pinches off to form an endosomal vesicle which is surrounded by a protein called clathrin. "Early" endosomes can pick up new proteins and other constituents as well as shed them as they move and mature through the cell. During this maturation process, protein pumps in the endosome lead to a decrease in the endosomal pH which can lead to conformation changes in protein structure and shedding of proteins. Eventually the "late" endosome reaches and fuses with the lysosome, an internal organelle which contains degradative enzymes. Undegraded components like viral nucleic acids or cholesterol, are delivered to the cell. This transport can also go in the reverse direction and recycle receptors to the cell membrane. Likewise vesicles pinched off from the Golgi complex can fuse with endosomes, with some components surviving the process to reenter the Golgi.
Synthesize/Degrade: Cells have to both synthesize and degrade small molecules as well as larger polymeric protein, carbohydrates and nucleic acids. The anabolic (synthetic) and catabolic (degradative) pathways are often compartmentalized in time and space within a cell. For example fatty acid synthesis is carried out in the cytoplasm but fatty acid oxidation is carried out in the mitochondria. Proteins are synthesized in the cytoplasm or completed in the endoplasmic reticulum (for membrane and exported proteins) while they are degraded in the lysosome or more importantly in a large multimolecular structure in the cell called the proteasome.
Let's consider some key characteristics of a cell before we get into the details.
Cells and their internal compartments have regulated concentrations of ions and hydronium ion.
As expected the pH of the cytosol (the aqueous substance surrounding all the organelles within the cell) varies from about 7.0-7.4, depending on the metabolic state of the cell. Some organelles have proton transporters that can significantly alter the pH inside an organelle. For example the pH inside the lysosome, a degradative organelle, is about 4.8. The collapse of the pH gradient across the inner mitochondrial membrane is sufficient to drive the thermodynamically unfavored synthesis of ATP.
Compared to the extracellular fluid, the concentration of potassium ion is higher inside the cell, while concentrations of sodium, chloride and calcium ions are higher on the outside of the cell (see table below). These concentration gradients are maintained by ion transporters and channels and require energy expenditure ultimately in the form of ATP hydrolysis. Changes in these concentrations are integral to the signaling system used by the cell to sense and respond to changes in its external and internal environments.
Water is found in bulk form as well as bound to macromolecules like proteins and polysaccharides. These waters would be expected to have different properties. The table below show approximate ion concentrations in the cell.
| Ion | Inside (mM) | Outside (mM) |
| Na+ | 140 | 5 |
| K+ | 12 | 140 |
| Cl- | 4 | 15 |
| Ca2+ | 1 uM | 2 |
Cells have an internal framework that provides architectural and internal structural support.
The "cytoskeletal" architecture of a (with molecular "cables"- and "girder-like" structures) is not dissimilar from a factory.
 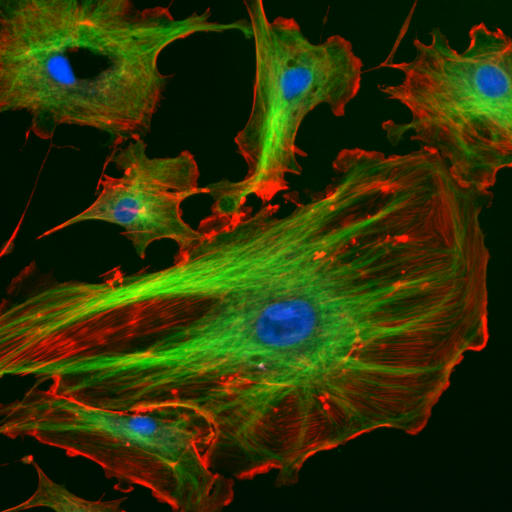 |
|
| http://www.cybercom.net/~copters/trips/pictures/factory_inside.jpg http://en.wikipedia.org/wiki/File:FluorescentCells.jpg |
The internal framework or cytoskeleton of a cell, is composed of microfilaments, intermediate filaments, and microtubules. These are comprised of monomeric proteins which self assemble to form the internal architecture. Parts of the cytoskeleton can be seen in the photo above (taken from Wikipedia).
Microfilaments of actin monomers (which are stained with a red/orange fluorophore) and microtubules which offer more structural support made of tubulin monomers (stained green) along with the blue-stained nucleus, are shown in the image. Organelles are supported and organized by the cytoskeleton (primarily microtubules). Even the cell membrane is supported underneath the inner leaflet by actin (stained orange) and spectrin microfilaments. Motor proteins like myosin (that moves along actin microfilaments) and dynein and kinesin (that move along tubulin microtubules) carry cargo (vesicles, organelles) in a directional fashion. The cell is not a disorganized collection of molecules and organelles. Rather is a highly organized for optimal chemical production, use and degradation.
Cell have a variety of shapes. Some circulating immune cells must slip through the cells that line capillary walls to migrate to sites of infection. The same process occurs when tumor cells metastasize and escape to other sites in the body. In order to do so, the cell must drastically change shape, a response that requires dissociation of the cytoskeleton polymers into monomers which are available later for repolymerization.
The cell is an amazingly crowded place.
In chemistry labs, we typically work with dilute solutions of solute molecules in a solvent. You have probably heard that the body is comprised of 68% water, but the water concentration is obviously dependent on the cellular environment. Solute molecules like protein and carbohydrates are densely packed. Cells are so crowded that the space between larger molecules like protein is less than the size of protein. Studies have shown that the stability of a protein is increased in such condition, which would help keep the protein in the folded, native state. Another consequence of high concentrations is that it would promote the binding or self aggregation of like molecules as well as dislike ones which from an equilibrium perspective would not occur in dilute solutions. Hence the study of biomolecules in dilute solutions in the lab may not reveal the actual complexities of interactions and activities of the same molecule in vivo. Recently investigators have added a neutral copolymer of sucrose and epichlorhydrine to cells in vitro. These particles induced organization of extracellular molecules secreted by cell, forming an organized extracellular "matrix" which induced the organization of the microfilaments on the inside of the cell as well as inducing changes in cell activity.1 In vitro enzyme activity of a key enzyme in glycolysis dramatically increases under crowded conditions.2 Another result of crowding may be the spatial and temporal association of key enzymes in specific metabolic pathways, allowing easier diffusion of substrates and products within the colocalized enzymes.
Figure: The crowded cytoplasm of E. Coli - Ufrom McGuffee SR, Elcock AH (2010) PLoS Comput Biol 6(3): e1000694. doi:10.1371/journal.pcbi.1000694 (open source journal)
The computer simulation used 50 different types of the most abundant macromolecules of the E. coli cytoplasm and 1008 individual molecules. Rendering of the cytoplasm model at the end of a dynamics simulation. RNA is shown as green and yellow. This figure was prepared with VMD
Cell components undergo phase transitions to form substructures within the cell.
A perplexing question is how do substructures form within a cell. This includes not only the biogenesis of organelles like mitochondria but also smaller particle such as polysaacharide granules, lipid droplets, protein/RNA particles (including the ribosome) as well as the nucleolus of the cell nucleus. It might be easiest to consider this problem using two examples from the lipid world, lipid droplets and membrane rafts. You are very familiar with phase transitions that occur when a sparing soluble nonpolar liquid is added to water. At a high enough concentration, the solubility of the nonpolar liquid is exceeded and a phase transition occur as evidenced by the appearance of two separate liquid phases. The same process occurs when triglycerides coalesce into lipid droplets with proteins associated on their outside. Another example occurs within a cell membrane when lipids with saturated alkyl chains self associate with membrane cholesterol (which contains a rigid planar ring system) to form a lipid raft characterized by greater packing efficiency, rigidity and thickness that other parts of the membrane. These lipid rafts often recruit protein involved in signaling processes within the cell membranes. This process of phase separation is also called liquid/liquid demixing as two "liquid-like" substances separate.
It appears that proteins that interact with RNA are composed of less diverse amino acid and have more flexible ("more liquid-like) structures allowing their preferential interaction with RNA to form large RNA-protein particles (like the ribosome and other RNA processing structures) in a fashion that mimics liquid/liquid demixing. All of these interactions are just manifestations of the various intermolecular forces you studied in earlier chemistry classes.
You have studied lipid, protein, nucleic acid and carbohydrate structure in introductory biology and/or chemistry classes. Now follow the following links which will take you to Cell Tutorial chapters that show the life history of these types of molecules in the cell.
A list of web sites and applets on eukaryotic cell structure:
-
Cell Tutorial from UCLA
-
Plant and animal cell simulation (requires Java)
-
Download Apple Store Applet for IPad, Itouch, IPhone: 3D Cell
References:
1. Zeiger AS, Loe FC, Li R, Raghunath M, Van Vliet KJ (2012) Macromolecular Crowding Directs Extracellular Matrix Organization and Mesenchymal Stem Cell Behavior. PLoS ONE 7(5): e37904. doi:10.1371/journal.pone.0037904 .
2. Apratim Dhara, Antonios Samiotakisb, Simon Ebbinghausa, Lea Nienhausa, Dirar Homouzb, Martin Gruebelea, and Margaret S. Cheungb. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl Acad Sci. www.pnas.org/cgi/doi/10.1073/pnas.1006760107
3. Anthony A. Hyman and Kai Simons. Beyond Oil and Water--Phase Transitions in Cells. Science 337, 1047 (2012); DOI: 10.1126/science.1223728
4. McGuffee SR, Elcock AH (2010) Diffusion, Crowding & Protein Stability in a Dynamic Molecular Model of the Bacterial Cytoplasm. PLoS Comput Biol 6(3): e1000694. doi:10.1371/journal.pcbi.1000694
This page was written and maintained by Henry V. Jakubowski, Ph.D., College of Saint Benedict / Saint John's University. These materials are available for educational use.
Send corrections to hjakubowski@csbsju.edu
This material is based upon work supported by the National Science Foundation under Grant No. 1043566.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Navigation
| Reactivity Index | Biochemistry Online: Table of Contents |
| Structure & Reactivity Web Materials |

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.