Biochemistry Online: An Approach Based on Chemical Logic

 Homework
Problems - Literature Learning Module: Carbohydrates 1
KEY
Homework
Problems - Literature Learning Module: Carbohydrates 1
KEY
Assessment of Biochemistry/Molecular Biology (BMB) Foundational Concepts
Henry Jakubowski, Ph.D., Professor, Chemistry Department, College of Saint Benedict/Saint John's University
Research Paper: Trypanosoma brucei Glycoproteins Contain Novel Giant Poly-N-acetyllactosamine Carbohydrate Chains.Abdelmadjid Atrih, Julia M. Richardson, Alan R. Prescott, and Michael A. J. Ferguson. J. Biol. Chem., Vol. 280, Issue 2, 865-871, January 14, 2005
A densely packed layer of a surface glycoprotein covers the surface of the African trypanosome, Trypanosoma brucei. The parasitic protozoa are transmitted by tsetse fly and causes African sleeping sickness in humans and nagana in livestock. In the mammalian host, T. brucei live and divide extracellularly in the blood, lymph, and interstitial fluids. They have 100's of genes for the glycoproteins which produce so many variants that the organism can elude the immune system. The parasite has a deep invagination of the plasma membrane, called the flagellar pocket. Endocytosis and secretion occurs from this site. Little is known about its unique membrane glycoprotein structures and the endosome (vesicles that have undergone endocytosis)/lysosome (degratory vesicles that fuse with lysosomes) system.. Something in the pocket binds the lectin ricin and tomato lectin. Ricin is a heterodimeric protein consisting of a Galβ1-4GlcNAc binding subunit, B chain (32 kDa), and a toxic subunit, A chain (ca. 30 kDa), connected through a disulfide linkage. Tomato lectin is known to bind a linear polymer of N-acetyllactosamine (poly-LacNac), which is comprised of up to 26 Galβ1-4GlcNAc linked principally by 4GlcNAcβ1-3Galβ1- links (which in mammals is found in both the cell and lysosomal membrane glycoproteins). A study was done to exam the flagellar pocket of the bloodstream form of Trypanosoma brucei by studying the ricin-binding glycoprotein.
1. Ricin-binding glycoproteins were purified as follows.
a. Cell were isolated from blood, subjected to hypotonic lysis to release cytosolic components and soluble forms of surface glycoproteins, and cell ghost (cells without cytoplasm and other soluble contents) collected. Briefly explain this procedure and how it works with words and pictures.
Ans: If deionized water is added to cells, water flows into the cells to balance ionic strength across the membrane, causing the cell to swell and burst. The mixture can be centrifuge. The supernatant contains soluble cell components, while the pellet contains membrane material referred to as ghosts.
b. Cell ghosts were solubilized in 50 mL of 8 M urea, 3% SDS, 50 mM Tris-HCl, pH 6.8. Explain the role of each of the added ingredients. How do they collectively solubilize the cell ghost.
Ans: Urea would denature any proteins. The SDS is an ionic detergent which also denatures proteins and would also solubilize the membrane lipids and any membrane protein in the ghost. All proteins would be denatured. The Tris-HCl is a buffer solution that keeps the pH constant at 6.8.
c. The extract was diluted 50 fold 50 mM Tris-HCl, pH 6.8, 0.4 M NaCl, 0.8% Triton X-100, and protease inhibitors. Ricin-coupled agarose chromatography beads were added and gently mixed overnight. The beads were collected by gentle centrifugation and packed into a colum. Why was the extract diluted 50 fold before the chromatography beads were added? Why did they switch from SDS to Triton X-100? Draw a diagram showing the ricin beads in the column and any interacting species.
Ans: On dilution, the concentration of the denaturing agents drops (urea from 8 M to 0.16 M and SDS from 3% to 0.06%) to levels that would probably not denature any proteins. This is important since in the next step they add a chromatography bead with the protein ricin coupled to it. If the denaturing agents were left at high concentrations, they would also denature the ricin, and prevent if from acting as a lectin and binding CHO on the solubilized ghost membrane proteins. They needed to add Triton X-100 since without some detergent, the membrane proteins solubilized in the previous step using SDS would come out of solution. Triton X-100 is a nonionic detergent that would form a micelle around the nonpolar parts of the membrane protein but does not denature membrane proteins as does SDS. Since SDS was added in the previous steps, the proteins would still be denatured but remain solubilized in a TX-100 micelle. Presumably the CHO on the membrane proteins would be able to bind to the protein ricin which is covalently attached to the chromatography bead.
d. Bound proteins were eluted from the column after extensive washing. Without altering pH or ionic strength, how could bounds proteins be eluted? Name the eluting agent.
Ans: Likely some sugar derivative with Gal . They used lactose and free Gal.
2. Aliquots of the eluted fractions were separated by PAGE. A membrane was placed on the gel and electrophoresis of the proteins onto the membrane was performed (Western Blot). The results are shown below.
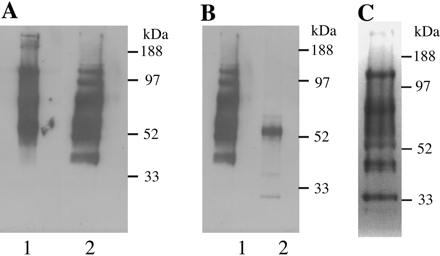
Fig 1. Ricin blot analysis of glycoproteins from bloodstream form T. brucei. Glycoproteins were eluted from a ricin-agarose column. After electrophoresis, glycoproteins were either transferred onto nitrocellulose membrane and probed with horseradish peroxidase-conjugated ricin (A and B) or stained for carbohydrate with periodic acid-Schiff (C). A: lane 1, native extract; lane 2, extract reduced with dithiothreitol (before electrophoresis). B: lane 1, reduced extract; lane 2, reduced extract after digestion with PNGase-F. C: reduced extract. The positions of molecular mass standards are indicated.
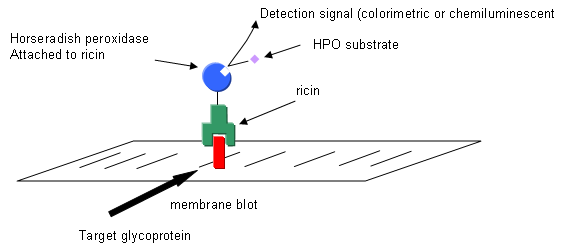
a. Draw a picture showing the electrophoreses protein on the membrane and how it react and is visualized with horseradish peroxidase-conjugated ricin
b. In gel A, explain the chemical effect of dithiothreitol on the proteins which do to alter the migration of the proteins.
Ans: dithiotreitol reduces disulfides in proteins. Proteins who mobility is unaffected to not have disulfides. Remember is disulfides remain in the protein after SDS treatment (which denatures proteins but does not cleave S-S bonds), the protein remains more compact that it would if it had the S-S bonds cleaved, causing it to migrate faster in the PAGE gel.
b. Why is there only one main band left after treatment with PNGase-F before the electrophoresis. (Hint: Google the enzyme).
Ans: Peptide:
N-Glycosidase F, also known as PNGase F, is an amidase that cleaves between the
innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex
oligosaccharides from N-linked glycoproteins
![]()
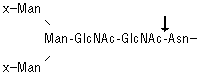
![]()
PNGase F hydrolyzes nearly all
types of N-glycan chains from glycopeptides/proteins. [x = H or sugar(s)]
PNGase-F digestion of the ricin-binding glycoproteins revealed that almost all of the ricin-binding glycans are N-linked structures (Fig. 1B). The remaining ricin binding band at about 50 kDa is most likely residual VSG, a proportion of which is known to express ricin-binding terminal β-galactosidase as a side chain on its glycosylphosphatidylinositol anchor.
3. Glycans released from the ricin-binding glycoproteins by PNGase-F were separated by Bio-Gel P4 gel filtration chromatography.
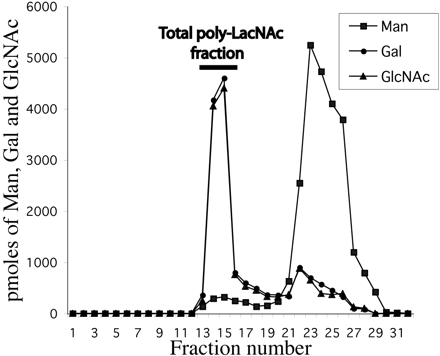
Fig. 2. Bio-Gel P-4 chromatogram of glycans obtained from ricin-binding glycoproteins. Glycans released from ricin-binding glycoproteins by the action of PNGase-F were purified from detergents and salts. The monosaccharide compositions of the fractions were determined by GC-MS. The void volume total poly-LacNAc fraction is indicated with a bar.
Two major fractions were found. Which peak has the highest average molecular weight species? Give an estimate of the molecular weight range of the glycans found in the void volume of the Bio-Gel P-4 column, based on the information from BioRad below.
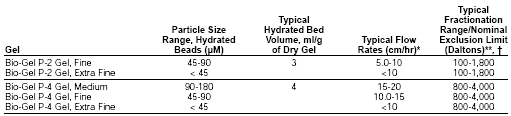
Ans: The first peak eluting in fractions 13-15 has the highest average MW. It comes through in the void volume. This peak would probably containing molecules with a range of molecular weights which don't separate from each other since they don't partition into the volume within the beads. The MW would be greater than 4000.
4. The poly-LacNAc fraction from the Bio-Gel P4 column was chromatographed on a HPLC Dionex CarboPac A-100 high pH anion exchange column. (HPAEC), used for the analysis of weakly ionizable carbohydrates. The beads in this column consist of 7.5-μm diameter vinylbenzyl chloride/divinylbenzene macroporous substrate fully functionalized with an alkyl quaternary ammonium group (15%crosslinked). The column is equilibrated in 95% solution A (100 mM NaOH) and 5% solution B (0.380 M sodium acetate in 0.1 M NaOH) and bound glycans separated using a linear gradient of 5-40% solution B.
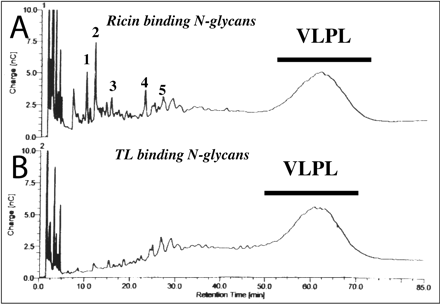
Fig. 3. Dionex HPAEC separation of total poly-LacNAc fractions. A, HPAEC chromatogram of glycans, detected by pulsed-amperiometric detection, of the total poly-LacNAc fraction isolated after Bio-Gel P-4 gel filtration (see Fig. 2). B, HPAEC chromatogram of the glycans of the total poly-LacNAc fraction which bind to tomato lectin-agarose. The VLPL (very large poly LacNac) is indicated with a bar.
What is the pH of the column during equilibration and running? Why was this column run at this pH.
Ans: If the two solutions are at 0.1 M NaOH, then pOH = 1, so pH = 13. The column is used for separation of weakly ionizable carbohydates. The high pH is required to pull protons from the sugar residues, allowing them to acquire negative charges. This then allows the CHO derivatives to be separated on an anion exchange column (since the CHOs are now negatively charged). Acetal links which hold monosaccharaides together in polysaccharides are resistant to base-catalyzed hydrolysis.
5. The VLPL fraction was subjected to monosaccharide compositional analysis by GC_MS. A 1 ul aliquots that was desalted and dissolved in water was mixed with 1 ul of acetonitrile with 2% formic acid and subjected to GC-MS. .It contained Man, Gal, and GlcNAc in the ration of 3:59:60. Is this consistent with the proposed glycan structure?
Ans: Yes since the ratio of Gal and GlcNAc are essentially identical.
6. It is necessary to know the linkage of the monosaccharide in the VLPL. Linkage analysis was done by converting glycans to partially methylated alditol acetates and analyzing them by GC-MS. The general chemistry involved in the formation of methylated alditol acetates used in methylation analysis is shown below. Definitive identification requires, in addition to the analysis of the MS pattern, the comparison of retention times with those of known standards
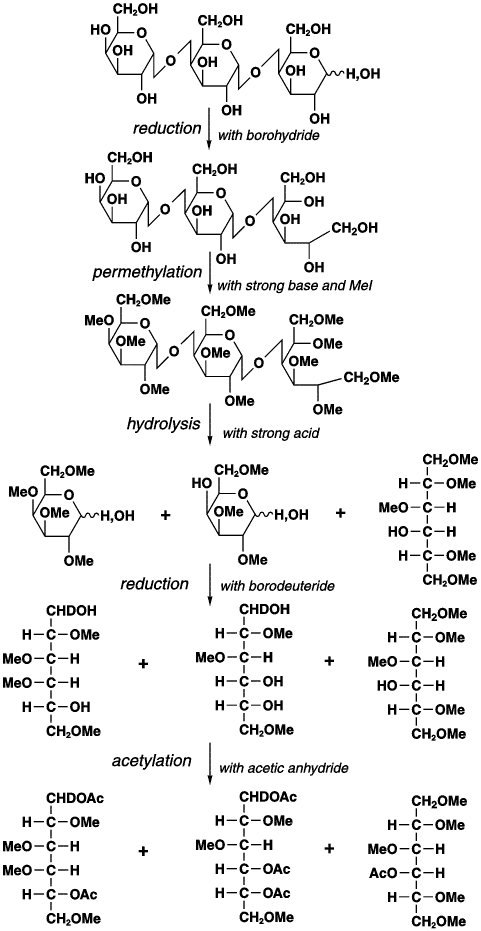
a. Why is methylation of free OH groups by basic CH3I required before hydolysis of the glycosidic links? What do methyl group denote? Ans: If the hydrolysis were carried out first, the OH group involved in the acetal link between monosaccharide residues would not be distinguished from other OH groups. The methylation serves to protect all OH groups not involved in the acetal link.
b. Why is the reduction carried out? Ans: Reductions causes the cyclic monosaccharaides, which are in equilibrium with the straight chain form aldehyde, to be convert completely to the straight form with an unmethylated OH residing on the C which was not protected by methylations and hence which was initially used to form the acetal link. The reducing agent acts on the straight chain aldehyde carbonyl C.
c. Why is a final acetylation carried out? What do acetyl groups mark? Ans: to complete the methylation of the free OH from the acetal link prior to MS.
"Traditional MS approaches are based on the fragmentation of organic molecules under some kind of impact, followed by the differentiation of the resulting particles according to their mass-to-charge ratio. In methylation analysis, peaks are identified by a combination of their retention time (relative time of elution from the LC column) and their electron impact (EI)-MS fragmentation pattern. The EI fragmentation patterns of similarly substituted monosaccharides of the same class (e.g., aldohexoses) are the same. Thus, definitive identification requires, in addition to the analysis of the MS pattern, the comparison of retention times with those of known standards (i.e., all 2,3,4-tri-O-methyl-hexoses produce the same EI-MS spectrum, but peracetylated 2,3,4-tri-O-methylgalactitol elutes later than peracetylated 2,3,4-tri-O-methylglucitol). This type of analysis indicates which residues are terminal (i.e., they are methylated at every position except OH-1 and OH-5) and how each monosaccharide is substituted; the latter reveals, inter alia, the occurrence of branching points. However, methylation analysis neither indicates which residues are attached to each other (i.e., does not provide sequence information) nor if a particular linkage is of the α or β configuration." NCBI
The methylation analysis in conjunction with NMR analysis showed large amounts of 4-)-substituted-(β)GlcNac and 6-O-substituted-(β)Gal. NMR analysis shows most of the GlcNAc and Gal residues were present as LacNac repeats of approximately 54.
Finally, fluorescein isothiocyanate-labeled ricin was added to fixed trypanosomes and detected by fluorescence microscopy.
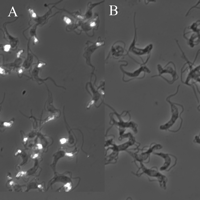
Fig 5. Subcellular localization of ricin-binding glycoproteins in bloodstream form T. brucei by fluorescence microscopy. Fixed trypanosomes were stained with FITC-ricin (A) and FITC-ricin in the presence of the ricin-blocking substances (as a control).
7. What ricin-blocking substances might they have used in panel B? Ans: galactose and lactose
It should be noted that the fluorescence indicates that ricin ligands are located mainly in the flagellar pocket and in the endosomal/lysosomal system of the trypanosome.
Navigation
Return to Biochemistry Online Table of Contents

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.