Biochemistry Online: An Approach Based on Chemical Logic

 Homework
Problems - Literature Learning Module: Lipids 1
KEY
Homework
Problems - Literature Learning Module: Lipids 1
KEY
Assessment of Biochemistry/Molecular Biology (BMB) Foundational Concepts
Henry Jakubowski, Ph.D., Professor, Chemistry Department, College of Saint Benedict/Saint John's University
Research Paper: Sphingomyelinase Activity Causes Transbilayer Lipid Translocation in Model and Cell Membranes. F. Xabier Contreras, Ana-Victoria Villar, Alicia Alonso, Richard N. Kolesnick and Félix M. Goñi (2003) The Journal of Biological Chemistry 278, 37169-37174 (doi: 10.1074/jbc.M303206200).
Sphingomyelin (SM) is a phospholipid localized mainly at the outer leaflet in microdomains or rafts of cell membranes. It can be cleaved by the protein enzyme sphingomyelinase, a phosphdiesterase, resulting in the generation of ceramide in the membrane. This enzyme cleaves SM in bilayer membranes.
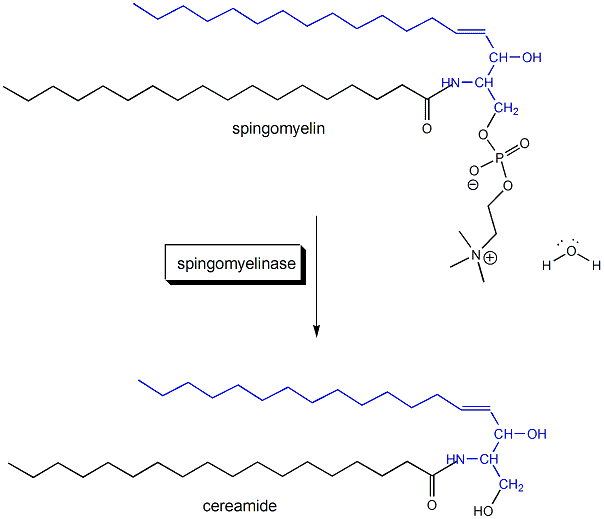
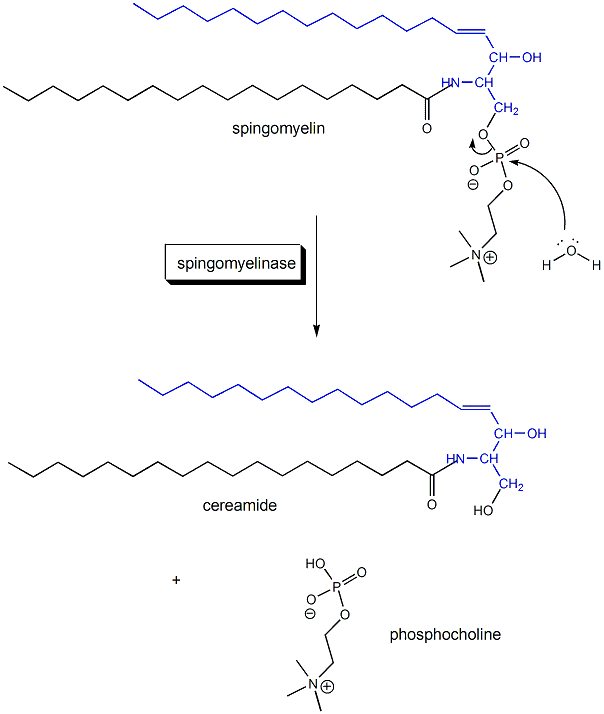
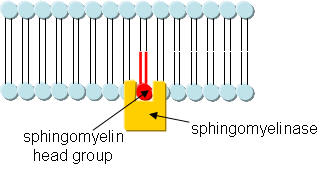
1. This reaction is a hydrolysis, catalyzed by the enzyme. Draw a mechanism showing the hydrolysis (in the absence of the enzyme, which would alter the mechanism) that would give ceramide. Draw the Lewis structure of the other product as well. Finally draw a cartoon of the enyzme with reactant in the active site.
Ans: O on H2O attacks the P in SM, forming ceramide and phosphocholine.
cartoon:
Sphingomyelinase (SMase) can be activated when an external signal, such as Vitamin D3 binds to a membrane receptor, initiating a signal transduction activation of the cell. One signal in the pathway would be alteration in the SM raft, leading to changes in membrane protein arrangement and activity. The study below was designed to address how SM hydrolysis alters lipid organization in bilayers.
2. LUVs were made with a lipid ratio of SM:PE:Cholesterol (2:1:1 molar ratio) and in the presence of a water soluble protein, neuraminidase (MW 70,000). Note that these are artificial liposomes not biological membranes. As such, the SM would be expected to be equally distributed between the two leaflets. Devise a method to remove free, unencapsulated neuraminidase from the LUVs.
Ans: Make liposomes using detergent dialysis method or by extrusion through membranes. Either way, neuraminidase would be found both encapsulated and outside of the membrane. Small MW molecules on the outside of the vesicles could be removed by dialysis since the vesicles in the dialysis bag would be too big to pass through pores in the bag. The easiest way would be to use gel fitration (size exclusion) chromatography, which could easily separate the relative huge liposomes (which would elute first) from the relatively small protein (which elutes later).
To study the properties of the LUVs, they were treated with GM3 ganglioside in methanol (in a volume 5% of the LUV preparation). A ganglioside is a glycolipid (contains a nonpolar lipid and polar sugar head group) usually found in membranes. The sugar head group of GM3 consists of neuraminic acid, galactose, and glucose, as shown in the structure below. Note that the encapsulated neuraminidase cleaves the terminal glycoyl-neuraminic acid from GM3. The most common types of gangliosides are glycosphingolipids. Since the final solution was 5% MeOH, an amount insufficient to alter liposome structure significantly, LUVs remained intact. Circle the polar head group.
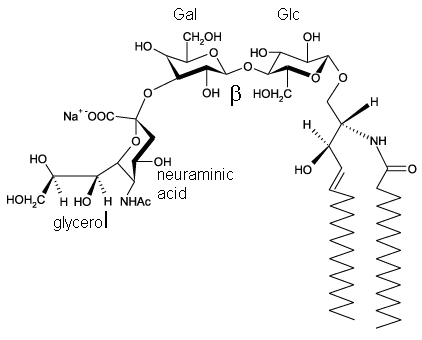
3. Draw a cartoon diagram showing both layers of the LUV before and after addition of GM3. Use simple geometric representations for the lipid and the protein neuramindiase. Don't use Lewis structures as cartoon representations for the lipids.
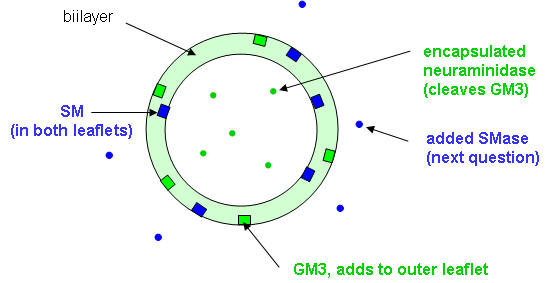
4. The water soluble enzyme sphingomyelinase (SMase) was added to a suspension of these vesicles. The extent of hydrolysis of as a function of time (from 0 to 60 minutes) and the effect on hydrolysis of excess amounts of the nonionic detergent Triton X-100 with time (60-90 minutes) are shown below in Figure 1A. Previous experiments had shown that this Triton X-100 concentration did not inhibit sphingomyelinase or neuraminidase.
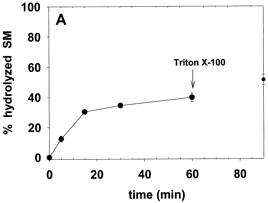
Figure 1A. Sphingomyelin hydrolysis by sphingomyelinase. Average values ± S.E. (n = 4). Average values ± S.E. (n = 5).
Explain using a cartoon and words the changes in the LUV on addition of SMase and of TX-100.
Ans: Addition of sphingomyelinase (1.6 units/ml) to a suspension of these vesicles induced SM hydrolysis, which reached equilibrium after 20 min, when 40% of SM had been hydrolyzed (Fig. 1A). Addition of Triton X-100 after 60 min caused membrane disruption, but SM hydrolysis did not go beyond 50% at 30 min after detergent addition. Suggest that SMase must be membrane bound to cleave SM. The 50% saturation level suggest that only SM in the outer leaftlet was cleaved, and not SM in the inner leaflet.
5. The same experiment was repeated (addition of SMase to the LUVs) but instead of monitoring SM hydrolysis, glycoyl-neuraminic acid cleavage from the added GM3 was studied. Aliquots of the vesicle suspension were removed at fixed times after the addition of sphingomyelinase and analyzed for the GM3 product of neuraminidase activity. The results are shown in Figure 1B below.
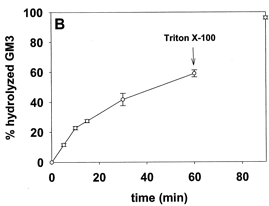
Figure 1B. GM3 ganglioside hydrolysis by entrapped neuraminidase. Average values ± S.E. (n = 5).
Describe the main difference in the results shown in graph 1b compared to 1A. Explain using a cartoon and words the changes in the LUV on addition of SMase and of TX-100.
Ans: GM3 was hydrolyzed almost in parallel with SM, except that no saturation was observed (Fig. 1B). After the addition of Triton X-100, virtually all GM3 was cleaved by neuraminidase, a water soluble enzyme that doesn't need an intact bilayer to cleave GM3. One interpretation of these data is that, as a consequence of sphingomyelinase activity, GM3 was flipping to the inner leaflet, thus becoming accessible to neuraminidase.
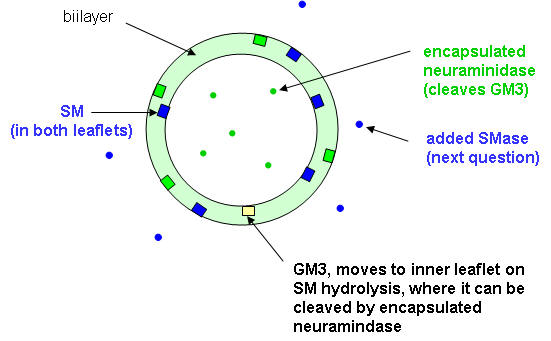
6. How was added GM3, which is found in the outer leaflet, cleaved by neuramidase, which is encapsulated? One explanation was that some encapsulated neuramidase leaked from the inside to the outside and the GM3 cleaved in the above experiment was in the outer leaftlet. To determine if neuraminidase was present on the outside of the LUVs, its activity against SMase treated vesicles as describe above was determined. The LUVs were filtered through membrane with small pores, allowing free neuramidase to pass through but not encapsulated LUV. They assayed the neuramidase using a soluble substrate of the enzyme, N-acetylneuraminyl-Gal-Glu. The results are shown in Figure 2A.
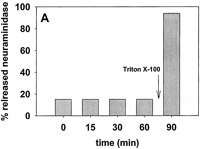
Figure 2A. Fraction of released neuraminidase in the time course of sphingomyelinase action. Free neuraminidase was separated from the vesicles by filtration. Average values ± S.E. (n = 3).
Does the constant presence of neuramindase present from 1-60 minutes suggest constant leakage from the LUVs or an leakage arising from the filtration method? (Note that the value at 0 minutes (the moment of SMase addition) is the same at later times. Explain. Are the results consistent with the cleavage of GM3 shown in figure 1B from leaked neuramindase?
Ans: It was necessary to rule out the possibility of neuraminidase coming out from the vesicles as a result of sphingomyelinase activity. To clarify this aspect, neuraminidase activity outside the sphingomyelinase-treated vesicles was assayed with the water-soluble substrate N-acetylneuraminyl-lactose. Even in the absence of sphingomyelinase activity (time zero), 18% of the total enzyme activity was recovered in the filtrates. This is probably because of vesicle breakdown due to shear stress during the filtration procedure used to isolate any "leaked" neuramidase from the encapsulated form.
7. Vesicles containing GM3 but not sphingomyelinase or internal neuraminidase were treated with neuraminidase at the concentration found in the filtrates above.
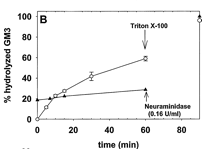
Figure 2B. ▲, hydrolyzed GM3 ganglioside when LUVs not containing neuraminidase were treated with the same amount of neuraminidase that was released in the experiment in 2A. Average values of two closely similar experiments; O, data of Fig. 1B (SMase added to LUVs with encapsulated neuraminidase) replotted for comparison.
Compare the times course of GM3 hydrolysis is totally different from the one depending on sphingomyelinase activity. Suggest a hypothesis to account for the enhanced hydrolysis of GM3 in the presence of SMase. Draw a cartoon diagram in your explanation.
Ans: Vesicles containing GM3 but not sphingomyelinase or internal neuraminidase were treated with neuraminidase at the concentration found in the filtrates. Neuraminidase was added to the vesicle suspension from the outside to ensure enzyme-substrate interaction. In this case (as seen in Fig. 2B), the time course of GM3 hydrolysis is totally different from the one depending on sphingomyelinase activity. Thus, the results in Fig. 1B cannot be explained on the basis of neuraminidase efflux from the vesicles.
8. In another experiment, antibody that binds and neutralizes the activity of neuraminidase was added to preformed LUVs and experiment 1 repeated. The graphs below the two cases
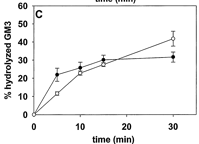
Figure 2C. GM3 ganglioside hydrolysis by LUVs with encapsulated neuraminidase, with and without antineuraminidase antibody added to the LUV suspension. Average values ± S.E. (n = 4). The paper did not define the symbols, but one is in the absence of added antibody and the other in the presence of added antibody. The curves differ little.
Using the data presented in Graphs 1-2, draw a cartoon and explain the sequence of events in the cleavage of GM3 by neuraminidase.
Ans: No significant differences were found between the fractions of hydrolyzed GM3 in the presence and absence of antibody. We concluded that GM3 hydrolysis was catalyzed by neuraminidase inside the vesicles; thus, sphingomyelinase activity had caused GM3 to flip to the inside lipid monolayer
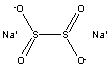
9. LUVs identical to those made above but with the added fluorophore, NBD-PE, were prepared. The structure of NBD-PE is shown below.
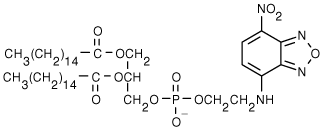
After the LUVs were made, the liposomes were treated with the membrane-impermeant sodium dithionite, which reduces NBD fluorescence.
Excess dithionite was removed immediately by gel filtration. Note: any remaining flourescence is from inner leaflet NBD-PE. LUVs were then treated with sphingomyelinase. Enzyme activity was the same as described in Fig. 1A. Aliquots were removed from the reaction mixture at regular intervals and mixed with an impermeant nonspecific antibody, IgG, labeled with another fluorophore, rhodamine. When NBD is excited with UV light, it fluorescence emission wavelength overlaps the excitation wavelength of rhodamine. If the two fluorphores are close enough, excitation of NBD can lead to rhodamine fluorescence emission, in a process called fluorescence resonance energy transfer.
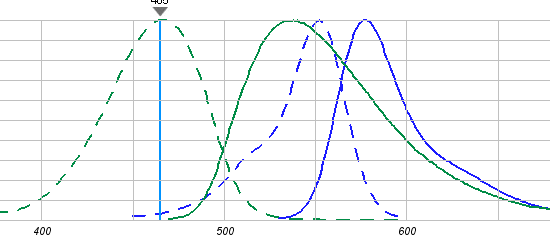
Figure: Excitation (---)/Emission (___)Spectra of NBD (green) and rhodamine (blue).
Results are shown in the Figure 3 below. Control experiments, also shown in the figure, indicated that in the absence of sphingomyelinase, NBD-PE fluorescence remained invariant with time.
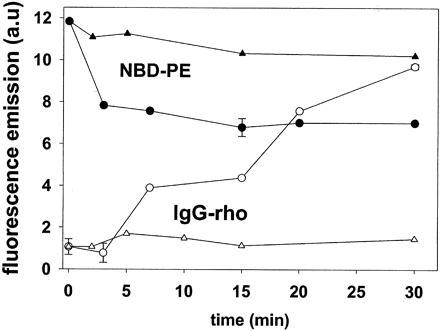
Figure 3. Sphingomyelin hydrolysis as in Fig. 1A. •, time course of NBD-PE fluorescence intensity; O, time course of Rho-IgG fluorescence intensity; triangles are the respective controls in the absence of sphingomyelinase.
Compare the relative changes see in the presence of SMase, with the relative lack of change in its absence. Are these results consistent with the results from the other experiments above?
Ans: As the sphingomyelinase reaction proceeded, NBD-PE flopped toward the outer monolayer, and energy transfer to Rho-IgG could take place. Consequently, when the fluorescence of NBD-PE was excited, its emission intensity decreased with time, and the emission intensity of Rho-IgG increased accordingly. The corresponding data are shown in Fig. 3. Control experiments, also shown in the figure, indicated that in the absence of sphingomyelinase, NBD-PE fluorescence remained invariant with time. Thus, fluorescence energy transfer measurements confirm that sphingomyelinase activity induces the transmembrane movement of lipids, in this case, the movement of NBD-PE from the inner to outer leaftlet.
Navigation
Return to Biochemistry Online Table of Contents

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.