Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
D: BINDING AND THE
CONTROL OF GENE
TRANSCRIPTION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/30/16
|
Learning Goals/Objectives for Chapter 5D: After class and this reading, students will be able to
|
D10. Epigenetic Control of DNA - Methylation
The fertilized egg is a totipotent cell. That is, through a series of divisions, its progeny cells can eventually become any of about 200 histologically different cell types. With subsequent cell division in the developing embryo, cells find themselves in different topological environments and have different cell-cell contact. Through signal transduction through the cell membrane, these cells start to become different, or differentiate, into other cells types. They do so by activating and inhibiting the expression of a different set of genes to form a different set of proteins in the cells. Most cells become terminally differentiated and eventually (after maybe a hundred cell divisions) lose the ability to divide and hence begin to die. However, a few types of cells, called stem cells, retain the ability to differentiate into other cells types in a regenerative process. These cells are pluripotent in that they can differentiate into other cell types.
How do dividing cells know what types of genes to actively transcribe? How can they have "memory" of the cells type they were before division? This appears to happen without alteration of the nucleotide sequence of the DNA in these cells. The main mechanism appears to be a inheritable but modifiable pattern of chemical modifications to the DNA (not unlike co- or post-translational modification of proteins) involving methylation/demethylation (by a methylase and demethylase) of cytosine in CpG dinucleotide repeats in the DNA. Also proteins can bind to DNA and methylated DNA to modify the course of gene expression in daughter cells. Such chemical modifications to the DNA which modify gene transcription are examples of epigenetic changes in the DNA.
You are all familiar with the cloning of animals from the DNA of adult cells (from Jurassic Park to the actual cloning of the sheep Dolly). In this process, the nucleus from an adult somatic cell (like a check epithelial cell) is removed and placed in an egg from which the nucleus has been removed (enucleated). The egg now has a full complement of DNA, just like an adult cell, but it didn't get the full set by normal means - i.e. by receiving half from a sperm to complement its own normal half. The DNA in the egg also has the methylation pattern of a terminally differentiated adult cell. It must be reprogrammed by undergoing extensive demethylation and remethylation to the "correct" epigenetic methylation state if the egg has a chance to form a normal embryo, fetus, and adult. Obviously this can happen, as evidenced by Dolly and success in cloning cows, cats, pigs, mice, and dogs. However, it is very difficult to achieve and probably accounts for the low success rate of cloning.
Methylation patterns can account for gene silencing (in which one gene in a pair of identical chromosomes is not expressed) and inactivation of one entire X chromosome in a female (who has 2 X chromosomes). In general, transcription from genes that are methylated is inhibited.
![]() Jmol: Updated
Methyl-CpG-Binding Domain of Human MBD1 in Complex with Methylated
DNA
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
Methyl-CpG-Binding Domain of Human MBD1 in Complex with Methylated
DNA
Jmol14 (Java) |
JSMol (HTML5)
Methylation patterns are inheritable and are also determined by the environmental variables. A recent study of neuron specific glucocorticoid receptor gene (NRGC1) promoter in the hippocampus showed an increase in methylation of the promoter and decreased levels of the mRNA transcript from the gene in suicide victims who had a history of childhood abuse compared to suicide victims that didn't suffer such abuse. These result paralleled those found in rats who were raised in a non-nurturing environment. The hippocampus is involved in the stress response.
Methylation patterns can be passed onto to daughter cells and on to offspring. This inheritance depends on specific binding of the DNA methylase to the methylated CpG on the template strand of the newly replicated DNA, which positions it to methylate the complementary newly replicated strand on the C hydrogen bonded to the template strand G. This enzyme specificity explains the requirement for a CpG dinucleotide methylation site in DNA, as shown below.
Figure: Methylation of CpG
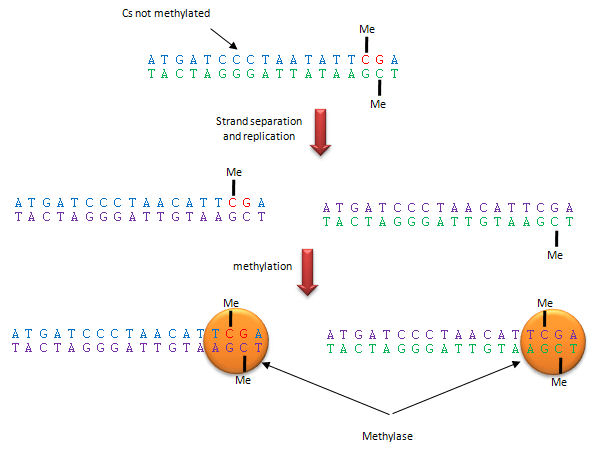
The dinucleotide sequence CpG is underrepresented in the human genome, yet is found in about 60% of promoter regions for genes. CpGs in promoter regions that are constitutively transcribed are not methylated. Methylated DNA outside of genes for proteins help to silence transcription from those regions. The CpG dinucleotide is probably found in low abundance since if 5 methyl cytosine spontaneously deaminates, it forms thymine which would lead from a CG to AT base pair mutation.
Yet another covalent epigentic modification has been discovered in neurons, where high levels of 5-hydroxymethyl cytosine are found. A 5-methylcytosine hydroxylase has also been discovered. Both probably have large roles in gene regulation.
Navigation
Return to Chapter 5D: Binding and the Control of Gene Transcription
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5D: Binding and the Control of Gene Transcription

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.