Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
D: BINDING AND THE
CONTROL OF GENE
TRANSCRIPTION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/30/16
|
Learning Goals/Objectives for Chapter 5D: After class and this reading, students will be able to
|
D3. Control of Gene Transcription in Eukaryotes
Three major differences exists in the control mechanisms used to regulate gene transcription in eukaryotes compared to prokaryotes.
-
multiple changes occur in the structure of chromatin at the site of transcription
-
positive mechanisms regulate transcriptions much more often than negative ones.
-
transcription and translation occur at spatially and temporally distinct sites and times.
The genomes of eukaryotes are much larger than prokaryotes. This poses some problems with respect to binding. Remember, DNA binding proteins demonstrate both nonspecific and specific binding. Nonspecific binding may help a protein find a specific site in the genome, but as the size of the genome increases, the chance of finding multiple specific sites randomly distributed increases. This problem can be avoided if multiple proteins are required to generate an active transcription complex. The chance of finding two or more specific sites for different proteins in proximity at sites other than required for gene transcription are very low. Multiple negative regulators would not be needed since just the binding of one regulator would probably be sufficient. Most eukaryotic genes have about 5 regulatory sites for binding transcription factors and RNA polymerase. Examples of these transcription factors are show in the figure below.
Figure:
![]() Control
of globin gene transcription
Control
of globin gene transcription
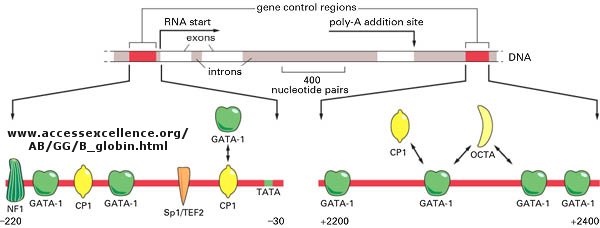
Figure:
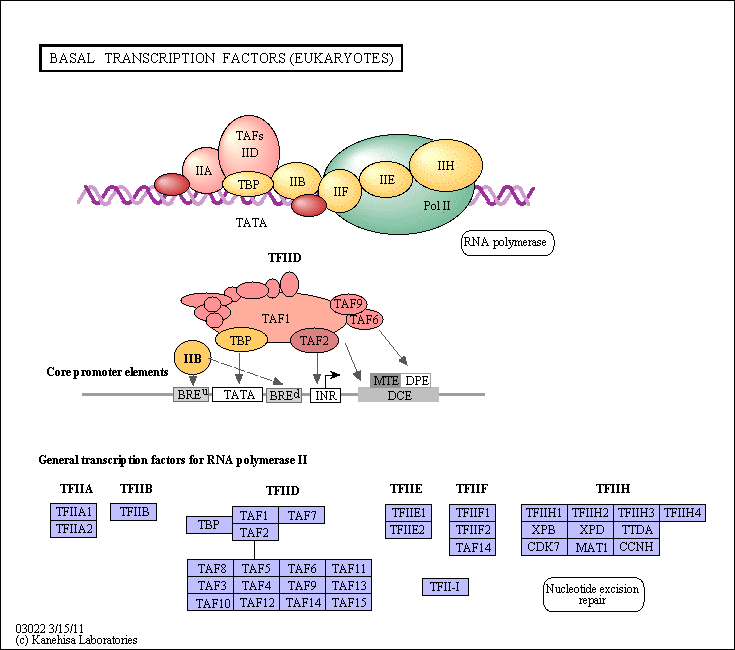
![]() Example
of transcription complexes
Example
of transcription complexes
(reprinted with permission from Kanehisa Laboratories and the KEGG project: www.kegg.org )
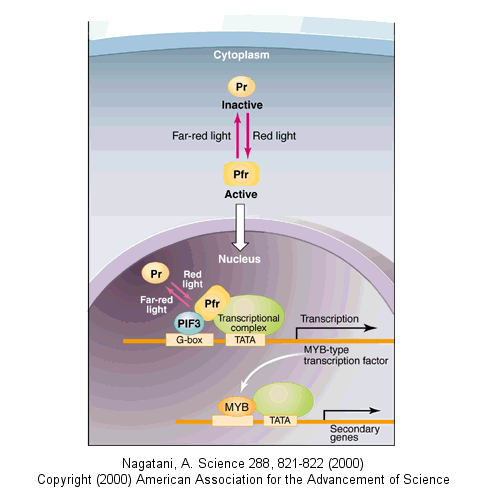
Light can even regulate gene expression by indirectly activating an inactive transcription factor in plants. The transcription factor PIF3 binds to a promoter region (G-box) of light-responsive genes. Only when PIF3 binds another transcription factor, Pr, is transcription activated. Pr, a "photoreceptor" is found in an inactive form in the cytoplasm. When it absorbs red light, it undergoes a conformational shift and moves into the nucleus, where it can bind PIF3 and activate transcription. The activation complex is inactivated when Pr interacts with far-red light.
![]() animation:
p53 and gene transcription from the HHMI (loads slowly)
animation:
p53 and gene transcription from the HHMI (loads slowly)
Navigation
Return to Chapter 5D: Binding and the Control of Gene Transcription
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5D: Binding and the Control of Gene Transcription

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.