Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
D: BINDING AND THE
CONTROL OF GENE
TRANSCRIPTION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/30/16
|
Learning Goals/Objectives for Chapter 5D: After class and this reading, students will be able to
|
D5. DNA Binding Proteins
Given the relative structural simplicity and repetitiveness of DNA, it would follow that proteins that bind specifically to it might have common DNA binding domain motifs but with specific amino acids side chains allowing for specific binding interactions.
- helix-turn-helix: found in prokaryotic DNA binding proteins.
Figure: helix-turn-helix
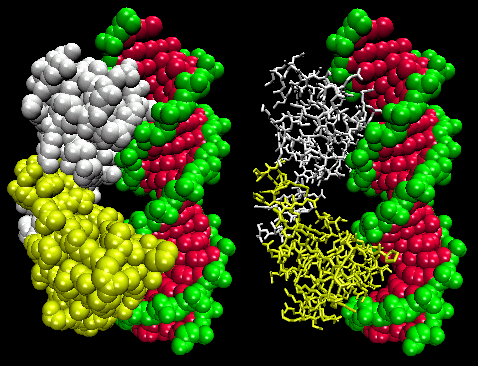
The figures shows two such proteins, the cro repressor from bacteriophage 434 and the lambda repressor from the bacteriophage lambda. (Bacteriophages are viruses that infect bacteia.) Notice how specificity is achieved, in part, by the formation of specific H-bonds between the protein and the major grove of the operator DNA.
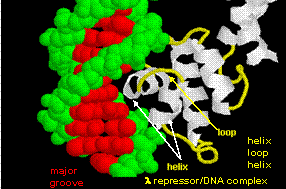
Figure: Lambda Repressor/DNA Complex
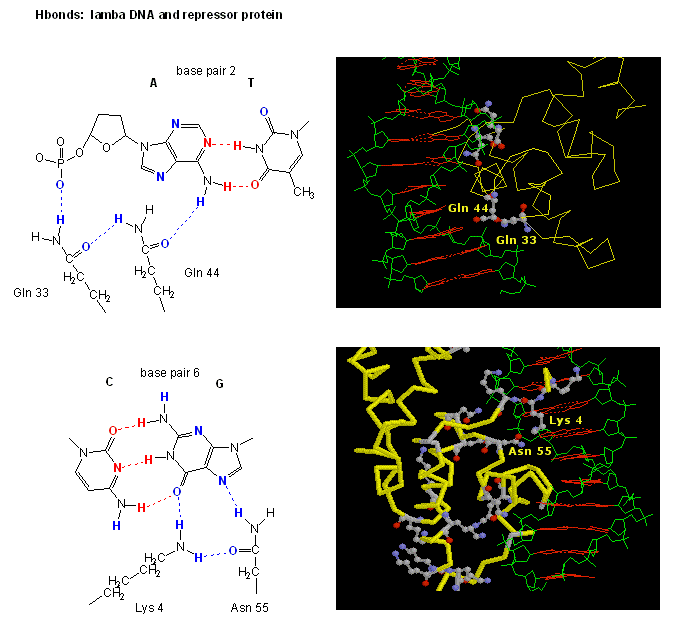
Figure: H Bond interactions betweenλ repressor and DNA
![]() Jmol:
Updated Lambda Repressor/DNA complex
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Lambda Repressor/DNA complex
Jmol14 (Java) |
JSMol (HTML5)
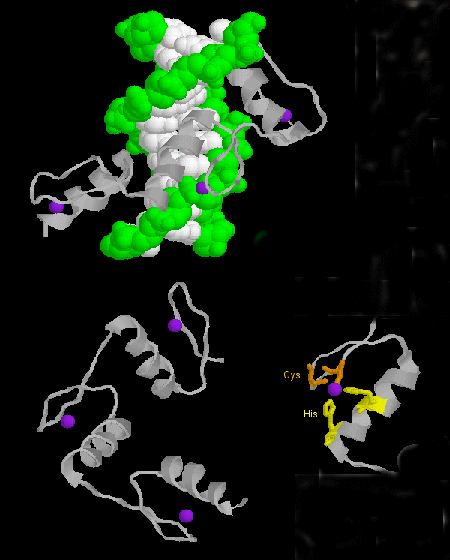
- zinc finger: (eukaryotes) These proteins have a common sequence
motif of
X3-Cys-X2-4-Cys-X12-His-X3-4-His-X4- in which X is any amino acid. Zn2+ is tetrahedrally coordinated with the Cys and His side chains, which are on one of two antiparallel beta strands, and an alpha helix, respectively. The zinc finger, stabilized by the zinc, binds to the major groove of DNA. ]
Figure: zinc finger
![]() Jmol:
Updated Zif268:DNA Complex
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Zif268:DNA Complex
Jmol14 (Java) |
JSMol (HTML5)
Zn finger proteins, of which 900 are encoded in the human genome (including the eukaryotic insulators binding protein CTCF described above) can be mobilized to actual repair specific mutations in cells, which if carried out in a high enough percentage of mutant cells could cure specific genetic diseases such as some forms of severe combined immunodeficiency disease. In this new technique (Urnov et al, 2005), multiple linked Zn finger binding domains, (one of the natural-occurring ones or mutant forms produced in the lab), each one specific for a certain nucleotide sequence, is linked to a nonspeciifc endonuclease, derived from the enzyme FokI. The nuclease is active in dimeric form so the active complex requires two endonuclease domains, each bound to four different Zn finger domains, to assemble at the target site. Specificity of binding is achieved by selection by the Zn finger domains. A nick is then made by the DNA by the nuclease, and host cell repair mechanisms ensue. This process involves strand separation, homologous recombination of the nicked region with complementary DNA within the cell, and repair of the nick. If excess wild type (non-mutated) DNA is added to the cells and uses as the template, the normal DNA repair mutation would fix the mutation. Urnov et al have shown the up to 20% of cultured cells containing a mutation can be repair in the lab. If these cells gain a selective growth advantage, the mutated cells would eventually be replaced with wild type cells.
- steroid hormone receptors: (eukaryotes) In contrast to most hormones, which bind to cell surface receptors, steroid hormones (derivatives of cholesterol) pass through the cell membrane and bind to cytoplasmic receptors through a hormone binding domain. This changes the shape of the receptor which then binds to a specific site on the DNA (hormone response element) though a DNA binding domain. In a structure analogous to the zinc finger, Zn 2+ is tetrahedrally coordinated to 4 Cys, in a globular-like structure which binds as a dimer to two identical, but reversed sequences of DNA (palindrome) within the major grove. (Examples of palindromes: Able was I ere I saw Elba; Dennis and Edna dine, said I, as Enid and Edna sinned.
Consider the glucocorticoid receptor (GR) as a specific example. It binds DNA as a dimer. The two DNA binding domains of the dimer associate with two adjacent major grooves of the DNA in the GR binding sequence (GBS), a short sequence of DNA within the promoter. Meijsing, et al. have found that not only does the GBS act as a binding site for GR, allowing transcription of genes, but it also affects the conformation of the receptor, causing gene transcription to be regulated in another way. The group constructed luciferase "reporters genes" which have GBS linked to the gene for the protein luciferase, that would express the protein luciferase (which fluoresces) if they were being transcribed, with the GBS. They found that relative transcriptional activity did not correlate to relative binding affinity of GR to the GBS. GBSs which were much more active than others bound comparably with those of lower activity, while GBSs with similar transcriptional activity bound with different affinities. This shows that the GBS is conferring unique function to the GR associated with it (i.e. transcription is not simply affected by whether or not the GR is bound to the GBS). A �lever arm� of the receptor was found to undergo conformational changes when bound to DNA, with changes specific to the sequence to which it was bound. A mutant protein, GR-γ, was made to be identical to the wild-type protein, GR-α, except in the lever arm was found to have different transcriptional activity even though they were binding to the same site on the DNA, showing that the lever arm and its conformation affects transcription.
- leucine zippers (or scissors): (eukaryotes) These proteins contain stretches of 35 amino acids in which Leu is found repeatedly at 7 amino acid intervals. These regions of the protein form amphiphilic helices, with Leu on one face, one Leu after two turns of a helix. Two of these proteins can form a dimer, stabilized by the binding of these nonpolar, leucine-rich amphiphilic helices to one another, forming a coiled-coil, much as in the muscle protein myosin. The leucine zipper represents the protein binding domain of the protein. The DNA binding domain is found in the first 30 N-terminal amino acids, which are basic and form an alpha helix when the protein binds to DNA. The leucine zipper then functions to bring two DNA binding proteins together, allowing the N-terminal bases helices to interact with the major grove of DNA in a base-specific fashion. Valine and isoleucine, along with leucine, are often found in stretches of amino acids that can interact to form other types of coiled coils.
Figure: leucine zippers (made with VMD)
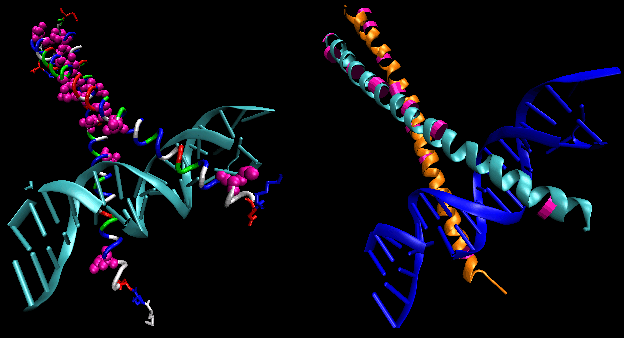
![]() Jmol: Updated
Leucine Zipper
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
Leucine Zipper
Jmol14 (Java) |
JSMol (HTML5)
Just as Zinc fingers nucleases have been used to induce repair of mutations, another study of the rat genome used specially designed ZFNs to cause breaks in ds-DNA that contain mutations from inaccurate DNA repair mechanism (by NHEJ) and hence contained specific mutations (Geurts, et al. 2009). This process, �knockout of the gene,� prevents the production of the protein normally transcribed by the target gene. Five- and six-finger ZFNs were used to achieve a high level of specificity in the targeted binding to the gene for three different proteins: green fluorescent protein (GFP), Immunoglobulin M (IgM) and Rab38. The knockout was successful in 12% of the rats tested; these animals had no wild-type protein and no expression. The ZFNs were sufficiently specific that no mutations were observed at any of 20 predicted non-target sites. This study supports the viability of control of transcription and expression for the treatment of disease and the importance of specific binding.
We have seen that two main factors contribute to the specific recognition of DNA by proteins; the formation of hydrogen bonds to specific nucleotide donors and acceptors in the major groove, and sequence-dependent deformations of the DNA helix to altered shapes with increased affinity of protein ligands. For example the Tata Binding Protein (TBP) can interact with a widened minor grove in the TATA box. New findings support that in addition proteins are able to use information in minor grooves that have become "narrowed" depending on the nucleotide sequence.
Tracks of DNA enriched in A can lead to twisting conformations that cause inter-base-pair hydrogen bonding in the major grooves, results in the narrowing of minor grooves. High amounts of AT base pairs are concentrated in narrow minor grooves (width <5.0 �) and CG base pairs are found more frequently in wide minor grooves.
How does minor groove narrowing affect DNA recognition? Narrow minor groves enhance the negative electrostatic potential of the DNA, making it a more specific and recognizable site. The backbone phosphates of the DNA are closer to the middle of the groove when it is narrow, thus correlating narrow minor grooves with a more negative electrostatic potential.
The minor grove-interacting parts of proteins contain arginine whose side chain can be accommodated into the more narrow and negative minor groove. Arginines can bind and in some cases insert themselves as short sequence motifs which enhance the specificity of the DNA shape recognition. Arg is preferred over Lys since the effective radii of the charge in Arg is greater than of the charge carrier in Lys. This would lead to a decreased desolvation energy for Arg which would promote its binding to the narrowed major grove. This discovery shows that "the role of DNA shape must be taken into consideration when annotating the entire genome and predicting transcription-factor-binding sites".
Figure: Arg in T3c Transposase binding in Narrowed Minor Grove of T3c Transposon
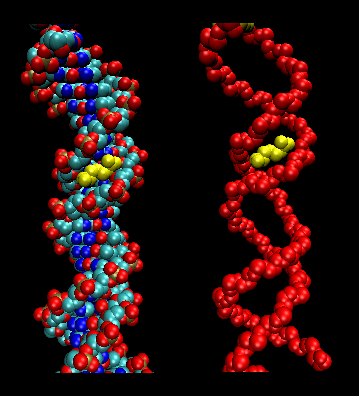
Sliding Model for protein/DNA Interactions
Navigation
Return to Chapter 5D: Binding and the Control of Gene Transcription
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5D: Binding and the Control of Gene Transcription

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.