Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
D: BINDING AND THE
CONTROL OF GENE
TRANSCRIPTION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/30/16
|
Learning Goals/Objectives for Chapter 5D: After class and this reading, students will be able to
|
D7. Phosphorylation and Control of Gene Expression
A common way to control gene expression is by controlling the post-translational phosphorylation of transcription factors by ATP. This modification might activate or inhibit the transcription factor in turning on gene expression. The added phosphate groups might be necessary for direct binding interactions leading to gene transcription or they might lead to a conformational change in the transcription factor, which could activate or inhibit gene transcription. A recent example of this later case is the control of the activity of the transcription factor p53. p53 has many activities in the cell, a primary one as a suppressor of tumor cell growth. It activates and inhibits a large number of genes in the process. If a cell is subjected to stress that results in genetic damage (an event which could lead a cell to transform into a tumor cell), this protein becomes an active transcription factor, leading to the expression of many genes, including those involved in programmed cell death and cell cycle regulation. Both of these effects could clearly inhibit cell proliferation. Hence p53 is a tumor suppressor gene. p53 is usually bound to the protein HDM2 which down regulates its activity by leading to its degradation. Stress signals lead to the activation of protein kinases in the cell (such as p38, JNK, and cdc2), causing phosphorylation of Ser 33 and 315 and Thr 81 in p53. This leads to the binding of Pin 1, a peptidyl-prolyl isomerase, which catalyzes the trans<=>cis conformational changes of X-Pro bounds. Pin 1 appears to bind only when p53 is phosphorylated. The ensuing change in p53 conformation presumably leads to its activation as a transcription factor.
Figure: Activation of p53 as a transcription factor by phosphorylation and conformational change
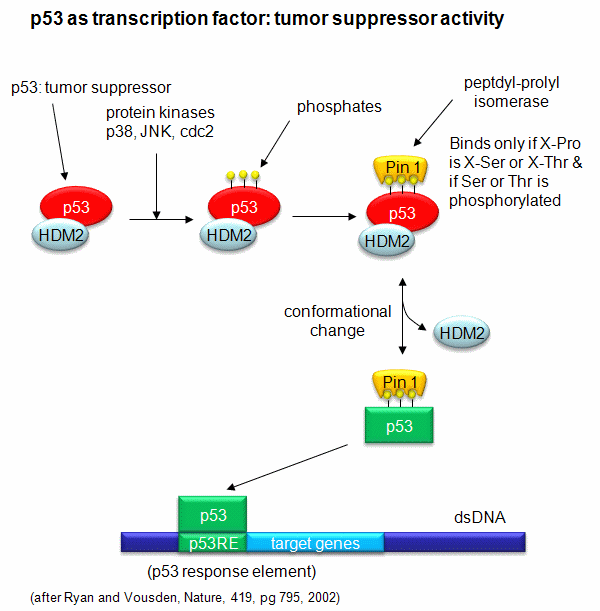
p53 binds to the p53 response element (p53RE) as shown above. In doing so it could either activate or repress gene transcription. The response element consists of two, 10-mers separated by a spacer of 0-13 nucleotides, and has the following canonical structure: RRRCWWGYYYspacerRRRCWWGYYY with W = A or T, R = C or G, and Y = C or T. The CWWG motif is required for high affinity binding of p53 to the response element. Alternations in the other nucleotides would fine tune binding and probably direct p53 to activate or repress transcription from the promoter. Over 1500 genes with a p53RE have been identified with many of these being repressed on binding of p53. Over 500 different high affinity binding sites for p53 have been found using microarrays. Wang et al have systematically altered the nucleotides in the p53RE linked to a promoter controlling the luciferase gene (which on activation and production of the protein luciferase can be monitored by light emission on addition of the substrate lucifein which releases a photon on reaction with dioxygen. They chose p53RE and promoters for two different genes, p21 whose transcription is known to be activated by p53, and Lasp1whose transcription is repressed. A figure representing the canonical sequence for both activating and repressing p53RE is shown below. (See the legend below for detailed explanation.) The dinucleotide sequence between the canonical C and G in each 10mer is a major determinant of whether p53 leads to activation or inhibition for transcription from promoter controlled by the response element.
Figure: Canonical sequence for both activating and repressing p53RE
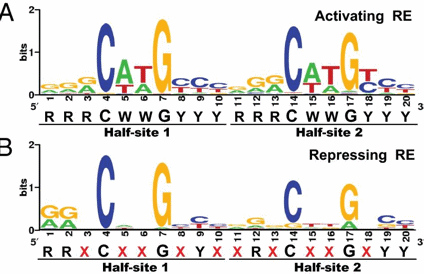
From Wang, Bei et al. Redefining the p53 response element. PNAS 2009 106 (34) 14373-14378; doi:10.1073/pnas.0903284106. Permission: Anyone may, without requesting permission, use original figures or tables published in PNAS for noncommercial and educational use (i.e., in a review article, in a book that is not for sale) provided that the original source and the applicable copyright notice are cited,
Each weblogo above (http://weblogo.berkeley.edu/logo.cgi ) consists of stacks of letters, one stack for each position in the sequence. The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding amino or nucleic acid at that position.
The repressive capabilities of the p53 protein have been studied quite extensively, yet little work seems to have been done surrounding what, if any, secondary mechanisms may affect the way it functions. The work of Wang et al attempts to address this problem. Through various substitutions of the base pair sequence they uncovered that two main elements seem to affect the function of this protein. First, the arrangement of a dinucleotide promoter directly affects whether or not p53 acts as a repressor or an activator. Some combinations will guarantee a certain function, whereas others depend more of the half-site background. Overall, the dinucleotide core seems to play an important role in the stabilization of the p53 to its response element. The second main way the function of the p53 can be modified is by the lateral triplet flank motifs. This implies that certain triplets of nucleotides located specifically in the base pair sequence can code for great activation. Determined by substitution of a �comprehensive set of synthetic triplet flanking motifs�, the RRR-YYY:RRR-YYY combination seemed to give a maximal activation. Moreover, they authors reviewed functions of previously reported p53 proteins. They found that some proteins thought to be activating were actually repressing, and some that were thought to be repressing, were actually activating. As a result, they insisted on paying greater attention to the base pair sequence, and avoiding deviations. After examining their study, it is clear that the p53 protein cannot continue to be defined as a single function protein. Rather, based on the evidence presented by the authors, the protein can act as either a repressor or an activator. This change of focus implies that it is necessary to reassess the previously analyzed p53 proteins to examine the true nature of their function.
Navigation
Return to Chapter 5D: Binding and the Control of Gene Transcription
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5D: Binding and the Control of Gene Transcription

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.