Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 7 - CATALYSIS
E: RIBOZYMES and the RNA World
BIOCHEMISTRY - DR. JAKUBOWSKI
06/10/2014
|
Learning Goals/Objectives for Chapter 7E: After class and this reading, students will be able to
|
"A foolish consistency is the hobgoblin of little minds...." Ralph Waldo Emerson
E1. Ribozymes
Any molecule that displays any of the catalytic motifs seen in the earlier chapters (general acid/base catalysis, electrostatic catalysis, nucleophilic catalysis, intramolecular catalysis, transition state stabilization) can be a catalyst. So far we have examined only protein catalysts. These can fold to form unique 3D structures which can have active sites with appropriate functional groups or nonprotein "cofactors" (metal ions, vitamin derivatives) that participate in catalysis. There is nothing special about the ability of proteins to do this. It is now known that RNA, which can form complicated secondary and tertiary structures as seen in the 3D image of the ribozyme from Tetrahymena thermophila, can as well.
Figure: ribozyme from Tetrahymena thermophila,

RNA molecules that act as enzymes are called ribozymes. This property of some RNA's was discovered by Sidney Altman and Thomas Czech, who were awarded the Nobel Prize in Chemistry in 1989. In contrast to protein enzymes which are true catalysts in that they are used over again, this is an example of a single use ribozyme. Other ribozymes are true catalysts and can carry out RNA slicing by transesterification (splicesome) and peptidyl transfer (in ribosomes). The mechanisms of catalysis of the hepatitis delta virus ribozyme include general acid/base catalysis.
Figure: mechanisms of catalysis
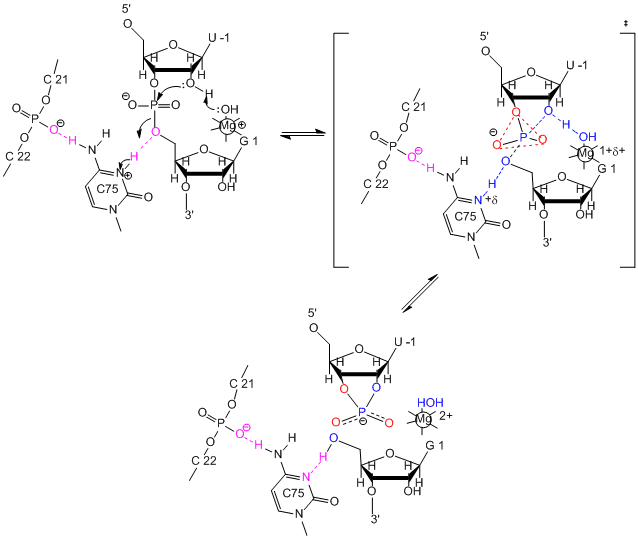
The hairpin ribozyme from satellite RNAs of plant viruses is 50 nucleotides long, and can cleave itself internally, or , in a truncated form, can cleave other RNA strands in a transesterification reaction. The structure consists of two domains, stem A required for binding (self or other RNA molecules) and stem B, required for catalysis. Self-cleavage in the hairpin ribozyme occurs in stem A between an A and G bases (which are splayed apart) when the 2' OH on the A attacks the phosphorous in the phosphodiester bond connecting A and G to form an pentavalent intermediate.
Figure: Self-cleavage in the hairpin ribozyme
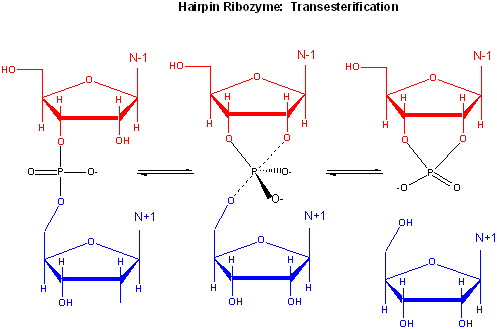
Rupert & Ferr�-D'Amar� (2001) solved the crystal structure of a hairpin ribozyme with a non-cleavable substrate analog containing a in which a 2'-OCH3 was substituted for the nucleophilic 2'-OH group. See the applet below.
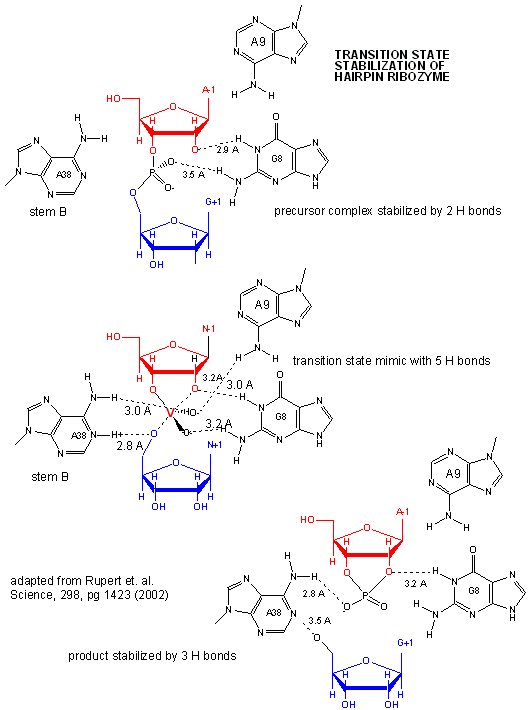
A recent study by Rupert et al. (2002) shows that A38 in Stem B appears to be able to interact with the products (the cleaved A now in the form of a cyclic phosphodiester with itself) and the departing G, and also with a transition state pentavalent analog of the sessile A-G bond in which the phosphodiester linking A and G in the substrate is replaced with a pentavalent vanadate bridge between A and G. However, A 38 does not appear to react with the sessile A -G groups in the normal substrate, indicating that the main mechanism used by this ribozyme is transition state binding. Since RNA molecules have fewer groups available for acid/base and electrostatic catalysis (compared to protein enzymes), ribozymes, presumably the earliest type of biological catalyst, probably make more use of transition state binding as their predominant mode of catalytic activity.
Figure: Active Site of Hairpin Ribozyme: Transition State Binding
Recently, the crystal structure of a purple bacterium group I self-splicing intron (which catalyze the removal of itself) interacting with both exons in a state prior to their ligation was determined (Adams, P. et al.). The structure shows both exons in close proximity. Nucleophilic attack of the 3'OH of the 5' exon on a distorted phosphate at the intron-3'-exon junction. Two metal ions reside on either side of the labile phosphodiester bond at the intron-3'exon junction, and are held in place by 6 phosphates.
A novel use of ribozymes was recently reported by Winkler et. al. They discovered that the 3' end of the mRNA of the gene glmS (from Gram-positive bacteria) which encodes an amidotransferase (catalyzing the formation of glucosamine-6-phosphate from glutamine and fructose-6-phosphate) is a ribozyme. A glucosamine-6-phosphate binding site in the ribozyme (3' end of the mRNA) binds this sugar, inducing autocleavage of the ribozyme. This inhibits, by an uncertain mechanism, the formation of the amidotransferase from the remaining part of the mRNA. This mechanism of regulation of gene expression through ribozyme activity might prove to be common.
In order for RNA molecules to have acted as both catalysts and carriers of genetic information in the original RNA world, RNA must have the potential to self replicate. Shechner et al. looked at the core structure of a class I ligase ribozyme able to polymerize RNA. This ligase was synthesized and enhanced through The tripod structure allows more RNA solvent interaction then often found in ribozymes. Several common structural motifs were also found to be present including the GNRA-triloop, uridine-turn and the frameshifting pseudoknot motifs. GNRA-triloop and uridine-turn motifs are short thermodynamically stable segments that cap the end of the helical regions. Varieties of these are found in many RNA structures although they don’t seem to be necessary for activity. The frameshifting pseudoknot motif is almost identical in structure to small viral ribosomal frameshifting pseudoknots and is adjacent to a new motif named “A-minor triad”. The A-minor triad is responsible for the coordination with Mg2+, although only when inserted in-between certain sequences. Triphosphoguanosine (called G1) at the 5’ terminus of the ribozyme acts as the electrophile for the RNA ligation reaction. Other segments of the ribozyme, such as the J1/2, have highly specific contacts which orient G1 and allow the RNA to fold more efficiently. Two residues, cytosine 12 (C12) and uracil 48 (U48) bind to Mg2+. Other sections of the RNA also promote specificity to assist in the replication process. A model for catalysis and the transition state of the ribozyme polymerase is similar to that for protein RNA polymerases, as shown in the figure below
Figure: Comparison of Transition State Models of Ribozyme and Protein RNA Polymerases (after Schechner et al)
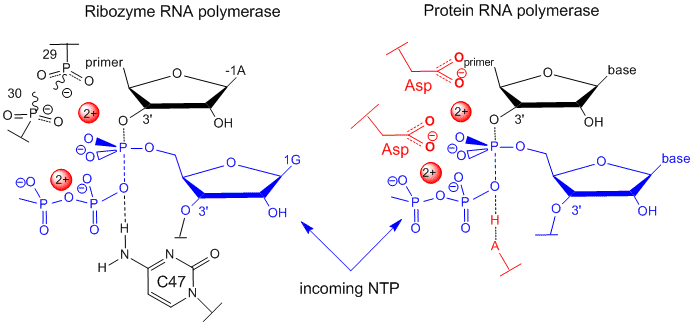
A divalent Mg2+ in the active site of the ribozyme enhances the nuclophilicity of the 3-OH on the primer, which attached the the terminal phosphate of the G(1)TP substrate to form a pentavalent intermediate. The Mg cation is stabilized by oxygens on P 29 and 30 of the ribozyme. The Mg ion also stabilizes the developing charge in the transition state and in the charge in the intermediate. Stabilization of analogous divalent cations in the protein polymerase occurs through Asp side changes in the protein.
- Hairpin Ribozyme Add 1HP6.pdb
![]() Jmol:
Updated Self-splicing Group I intron with both exons
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Self-splicing Group I intron with both exons
Jmol14 (Java) |
JSMol (HTML5)
![]() Jmol:
Updated L1 Ligase Ribozyme
Jmol14 (Java) |
JSMol (HTML5) Not done; Fix
Jmol:
Updated L1 Ligase Ribozyme
Jmol14 (Java) |
JSMol (HTML5) Not done; Fix
-
 3D
Structure of Ribozyme with Active Site - From the Howard Hughes
Medical Institute.
3D
Structure of Ribozyme with Active Site - From the Howard Hughes
Medical Institute.
Navigation
Return to 7E: Ribosomes and the RNA World Contents
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 7E: Ribozymes and the RNA World

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.