Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 7 - CATALYSIS
E: RIBOZYMES and the RNA World
BIOCHEMISTRY - DR. JAKUBOWSKI
06/10/2014
|
Learning Goals/Objectives for Chapter 7E: After class and this reading, students will be able to
|
"A foolish consistency is the hobgoblin of little minds...." Ralph Waldo Emerson
E3. The RNA World
Given that RNA expresses catalytic activities and can carry genetic information (some viruses have ds and ss RNA as their genome), it has been suggested that early life might have been based on RNA. DNA would evolve later as a more secure carrier of genetic information. An inspection of chemical properties of DNA, RNA, and proteins shows them to have attributes needed for their expressed function. Let's examine each for structural features that might be important for function.
a. Why does DNA lack a 2' OH group (found in RNA), which has been replaced with a hydrogen? This required the evolutionary creation of a new enzyme, ribonucleotide reductase, to catalyze the replacement of the OH in a ribonucleotide monomer to form the deoxyribonucleotide form. One possible explanation if offered in the figure below. DNA, the main carrier of genetic information, must be an extremely stable molecules. An OH present on C'2 could act as a nucleophile and attack the proximal P in the phosphodiester bond, leading to a nucleophilic substitution reaction and potential cleavage of the link. RNA, an intermediary molecule, whose concentration (at least as mRNA) should rise and fall based on the need for a potential transcript, should be more labile to such hydrolysis.
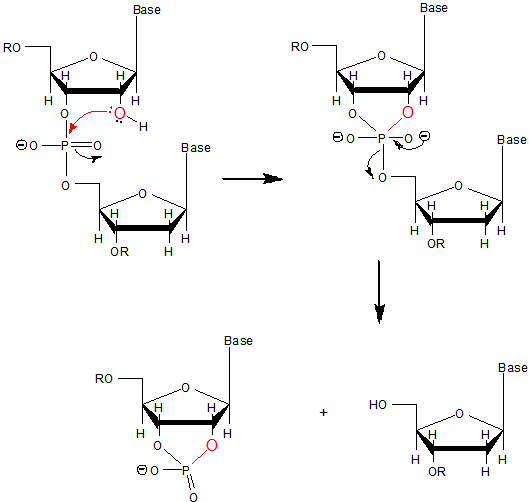
b. Why do both DNA and RNA contain a phosphodiester link between adjacent monomers instead of more "traditional" links such as carboxylic acid esters, amides, or anhydrides? One possible explanation is given below. Nucleophilic attack on the sp3 hybridized P in a phosphodiester is much more difficult than for a more open sp2 hybridized carboxylic acid derivative. In addition, the negative charge on the O in the phosphodiester link would decrease the likelihood of a nucleophilic attack. The negative charges on both strands in ds-DNA probably helps keep the strands separated allowing the traditional base pairing and double stranded helical structure observed.
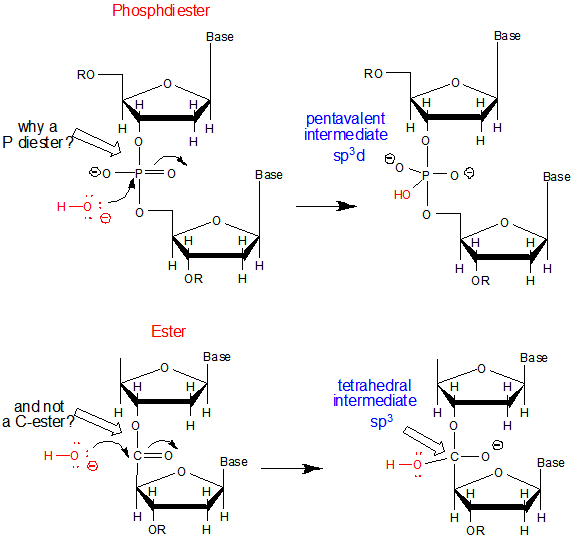
c. Why is DNA found as a repetitive double-stranded helix but RNA is usually found as a single stranded molecule which can form complicated tertiary structures with some ds-RNA motifs?
Another reason for the absence of the 2' OH in DNA is that it allows the deoxyribose ring in DNA to pucker in just the right way to sterically allow extended ds-DNA helices (B type). The pucker in deoxyribose and ribose can be visualized by visualizing a single plane in the sugar ring defined by the ring atoms C1', O and C4'. If a ring atom is pointing in the same direction as the C4'-C5' bond, the ring atom is defined as endo. If it is pointing in the opposite direction, it is defined as exo (see Jmol and figure below). In the most common form of double-stranded DNA, B-DNA, which is the iconic extended double helix you know so well, C2' is in the endo form. It can also adopt the C3' endo form, leading to the formation of another less common helix, more open ds-A helix. In contrast, steric interference prevents ribose in RNA from adopting the 2'endo conformation, and allows only the 3'endo form, precluding the occurrences of extended ds-B-RNA helices but allowing more open, A-type helices.
![]() Jmol:
Puckering in ribose and deoxyribose
Jmol:
Puckering in ribose and deoxyribose
![]() Jsmol:
Jsmol:
![]() Comparison
of ds DNA forms
Comparison
of ds DNA forms
![]() Jmol:
Comparison of ds DNA and RNA
Jmol:
Comparison of ds DNA and RNA
The figure below shows another comparison between the A-RNA and B-DNA double helices
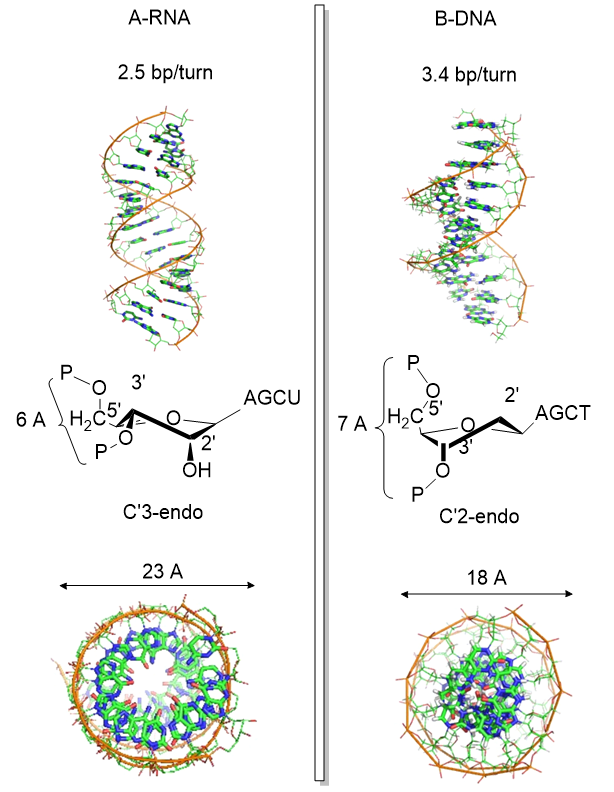
after Zhou et al Nature Structural and Molecular Biology. doi:10.1038/nsmb.3270
d. What about the molecular dynamics of A-RNA and B-DNA?
The information above suggests that the sugar ring of DNA is conformationally more flexible than the ribose ring of RNA. This can clearly be inferred from the observation that dsDNA can adopt B and A forms, which requires a switch from the 2' endo in the B form to the 3'endo form in the A form. The smaller H on the 2'C would offer less steric interference with such flexibility. The rigidity in ribose is associated with a smaller 5'O to 3'O distance in RNA leading to a compression of the nucleotides into a helix with a smaller number of base pairs/turn.
The increased flexibility in DNA allows rotation around the C1'-N glycosidic bond connecting the deoxyribose and base in DNA, allowing different orientations of AT and GC base pairs with each other. The normal "anti" orientation allows "Watson-Crick" (WC) base pairing between AT and GC base pairs while the altered rotation allows "Hoogsteen" (Hoog) base pairs. The different orientations for an AT base pair are shown below.
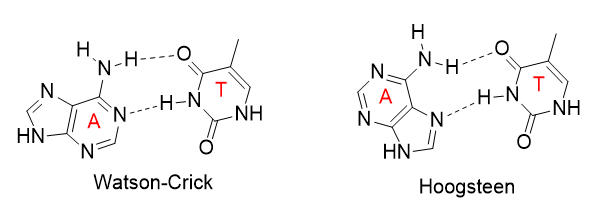
Hoogsteen base pairs can be found in distorted dsDNA structures (caused by protein:DNA interactions) but also in normal B-DNA. The figures below shown Watson-Crick and Hoogsteen AT base pairs in the MATa2 homeodomain:DNA complex (pdb 1K61). Note that the dA base in the Hoogsteen base pair is rotated syn (with respect to the deoxyribose ring) instead of the usual anti, allowing the Hoogsteen base pair.
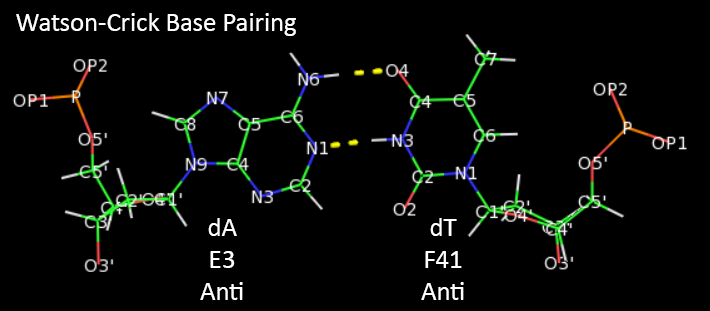
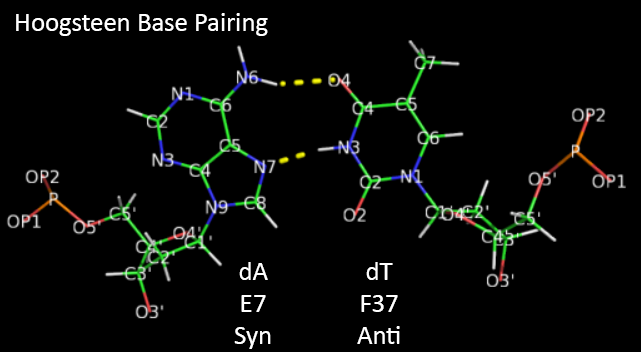
Studies (Zhou et al, 2016) show that Watson-Crick (WC) and Hoogsteen (HG) base pairs in B-DNA are in a dynamic equilibrium with the equilibrium greatly favoring the WC form. In a DNA:protein complex, the WC <----> HG equilibrium can actually favor the WG form for AT and GC+ forms (in the latter, the C is protonated) when those base pairs are also involved in protein recognition. They can also occur more frequently in damaged DNA. In contrast, molecular dynamic studies show that the HG base pairs A-U and GC+ are strongly disfavored in ds A-RNA.
One type of DNA damage is methylation on N1-adenosine and N1-guanosine. This modification prevents normal Watson-Crick base pairing but for DNA, these modified bases can still engage in Hoogsteen base pairing, preserving the overall structure of dsDNA and its ability to stably carry genetic information. This same methylation occur normally in post-transcriptional modified RNA. Hence, N1 adenosine and N1 guanosine methylation prevents any type of base pairing in the modified RNA. These properties make DNA a better carrier of molecular information and offers another way to regulate the structural and functional properties of RNA.
A Structural Comparision
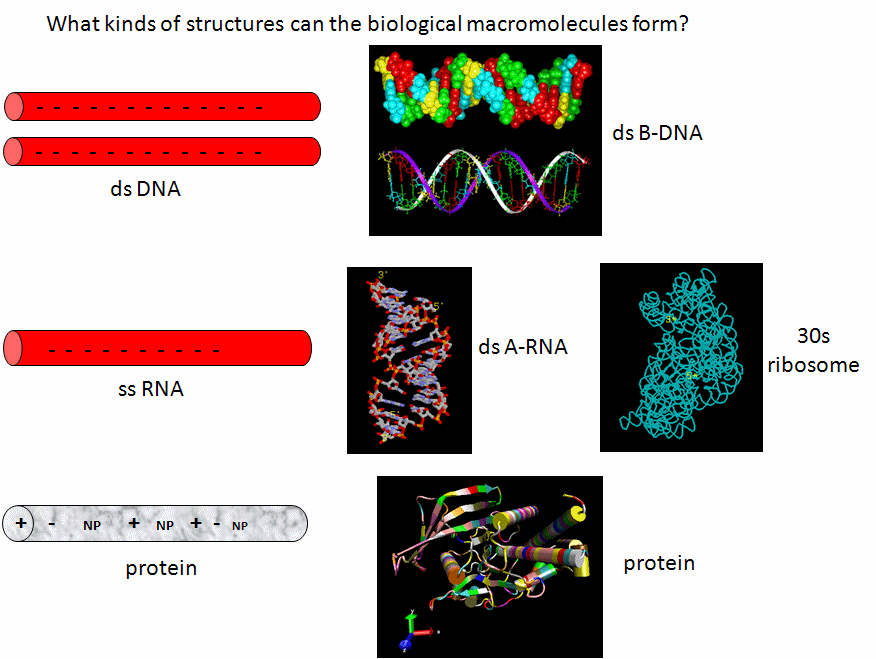
Now lets review the kinds of structure adopted by the 3 major macromolecules, DNA, RNA and proteins. DNA predominately adopts the classic ds-BDNA structure, although this structure is wound around nucleosomes and "supercoiled" in cells since it must be packed into the nucleus. This extended helical form arise in part from the significant electrostatic repulsions of two strands of this polyanions (even in the presence of counterions). Given its high charge density, it is not surprising that it is complexed with positive proteins and does not adopt complex tertiary structures. RNA, on the other hand, can not form long B-type double-stranded helices (due to steric constraints of the 2'OH and the resulting 3'endo ribose pucker). Rather it can adopt complex tertiary conformations (albeit with significant counterion binding to stabilize the structure) and in doing so can form regions of secondary structure (ds-A RNA) in the form of stem/hairpin forms. Proteins, with their combination of polar charged, polar uncharged, and nonpolar side chains have little electrostatic hindrance in the adoption of secondary and tertiary structures. That RNA and proteins can both adopt tertiary structures with potential binding and catalytic sites makes them ideal catalysts for chemical reactions. RNA, given its 4 nucleotide motif can clearly also carry genetic information, making it an ideal candidate for the first evolved macromolecules enabling the development of life. Proteins with a great abundance of organic functionalities would eventually supplant RNA as a better choice for life's catalyst. DNA, with its greater stability, would supplant RNA as the choice for the main carrier of genetic information.
Navigation
Return to 7E: Ribosomes and the RNA World Contents
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 7E: Ribozymes and the RNA World

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.