01/27/14
|
Learning Goals/Objectives for Chapter 3B: After class and this reading, students will be able to
|
This guide on carbohydrates will review those features that are deemed especially important for a one semester Biochemistry course dealing with structure/function relationships of biological molecules.
D. Glycoprotein Biosynthesis
Carbohydrates are added to proteins in a very complicated process which involves two organelles, the endoplasmic reticulum and the Golgi apparatus. CHO addition to proteins occurs both co- and post-translationally. The RNA coding the protein sequence enters the cytoplasm where it binds to ribosomes (large RNA-protein complexes) which are the site for protein synthesis. Cytoplasmic proteins are synthesized on free ribosomes, but for future glycoproteins, the ribosomes bind to an elongated, extensive organelle in the cell called the endoplasmic reticulum (ER).
![]() Figure: ribosomes
bind to an elongated, extensive organelle in the cell called the endoplasmic
reticulum (ER)
Figure: ribosomes
bind to an elongated, extensive organelle in the cell called the endoplasmic
reticulum (ER)
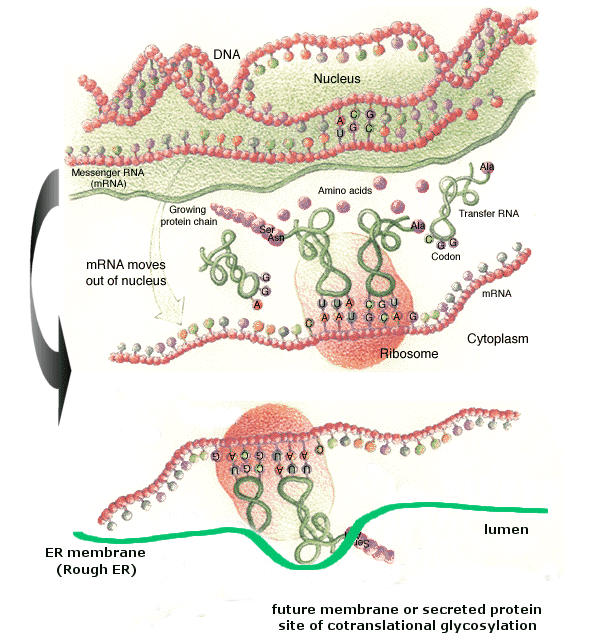
(from web, searching for reference; note: anticodon mislabeled as codon)
The nascent protein chain enters the lumen of the ER and a core oligosaccharide is added to the protein. Further additions and removal (trimming and processing) of monosaccharides are preformed in the lumen until a final core mannose structure has been added. This is demonstrated in the animation below. (The pink two-lobbed structure represents the ribosome which is bound to the ER membrane. The newly synthesized protein is shown in the lumen side of the ER membrane. That is where glycan transfer occurs.) The ER lumen contains high concentrations of molecular chaperones to assist protein folding.
Now additional carbohydrate modifications (post-translational) are made as the protein moves from the lumen of the ER (probably by a budding process) to another series of stacked, pancake-like organelles called the Golgi apparatus. Here terminal carbohydrate modification is completed. The Golgi does not contain molecular chaperons since protein folding is complete when the proteins arrive. Rather they have high concentrations of membrane bound enzymes, including glycosidases, and glycosyltransferases.
B. Glyprotein Function
The role of CHO in glycoprotein structure/function is slowly being determined. The most important seems to involve their role in directing proper folding of proteins in the ER which accounts for the observations that glycan addition to proteins in the ER is a cotranslational event. When inhibitors of ER glycosylation are added to cells, protein misfolding and aggregation are observed. The extent of misfolding depends on the particular protein and particular glycosylation sites with the protein. The polar CHO residues help promote solubility of folding intermediates, similar to the effects of many chaperone proteins.
The glycan moieties of the folding glycoprotein also lead to binding of the protein to lectins in the ER which serve as molecular chaperones. The most studied of these chaperones are involved in the calnexin-calreticulin cycle, and facilitate correct disulfide bond formation in the protein. After two glucose residues are removed by glucosidase I and II, the monoglucosylated protein binds to calnexin (CNX) and/or calreticulin (CRT), two homologous ER lectins specific for monoglucosylated proteins. Once bound, another protein, ERp57, a molecular chaperone with a disulfide bond (shown in diagram) interacts with the protein. This protein has protein disulfide isomerase activity.
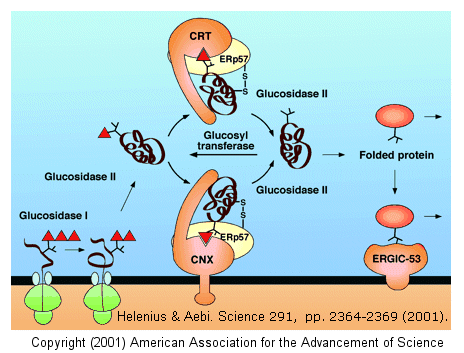
If a glycoprotein has not folded completely, it is recognized by a glycoprotein glucosyltransferase, which adds a glucose to it. This then promotes reentry into the calnexin/calreticulin cycle.
Ideally, unfolded or misfolded proteins would be targeted from degradation and elimination from cells. The ER has evolved a system to accomplish this. Since folding occurs in the ER, to prevent misfolding and aggregation, the ER also contains chaperones and folding catalysts. Stress (such as through heat shock) stimulates ER chaperone activity. As a final defense mechanism, unfolded or aberrantly-folded proteins are degraded by the cytoplasmic proteasome complex. Nonnative forms of some proteins that "escape" this surveillance system can accumulate and result in disease (for example neurodegenerative diseases like Alzheimers and Parkinson's disease.
C. Glycobiology and Glycomics
Our understanding of the synthesis and structure of glycan portions of glycoproteins has lagged behind our understanding of protein and nucleic acid structure and synthesis. Several reasons account for this:
New techniques in analysis and synthesis of carbohydrate analogs and inhibitors of enzymes involved in CHO synthesis and degradation, as well as in genetic manipulations of gene for these enzymes, are revolutionizing our understanding of the function of carbohydrate groups on lipids and proteins. On a more practical note, new methods to synthesize glycoproteins using recombinant DNA technology have been developed that allow synthesis of therapeutic human glycoproteins in yeast. Although they share many of the same synthetic steps, yeast glycoproteins are enriched n the high-mannose type, making them targets of the human immune system. Human and yeast glycoproteins synthesis produce the same mannose core in the ER (as illustrated in the animation above). However, differences in synthesis occurs in the Golgi. Human Golgi contain α-mannosidases I and II, which remove all but 3 Man residues from the final product. In yeast, however, these mannosidases appear to be missing so more Man residues are added (as many as 100). Human proteins made in yeast, therefore, contain many Man residue, which are recognized by the human immune system. So address these issues, Hamilton et al produced mutants of the yeast Pichia pastoris that localized glycoprotein synthesis proteins for mannosidase I and II, as well as other human glycoprotein synthesis genes, to the correct intracellular location while inactivating normal yeast gene, resulting in the production of human glycoproteins with the correct CHO structure in yeast. This may prove to have widespread use in the production of therapeutic human proteins.
May et al conducted experiments to determine the mechanism that allows enzymes to synthesize carbohydrate polymers of a defined length without a template (as in DNA, RNA, and protein synthesis). In their experiments they used GlfT2, a galactofuranosyltransferase which catalyzes the formation of galactan, a polymer of 20-40 galactofuranose (Gal f) residues that is an integral part of the cell wall in mycobacteria.. They first looked to see if the enzyme could regulate length by itself. To do this they used only the GlfT2 and the sugar with a synthetic lipid base replacing the natural one. The experiment showed that he GlfT2 could make galactan of length with a peak at 27 monomers by itself. They then wanted to know if GlfT2 used a processive process (when the transferase releases the growing glycan after many rounds of addition of monomer) or a distributive process (when the transferase releases the growing glycan after each monomer addition). To do this they added a reagent that prevented the GlfT2 from rebinding to a growing polymer after its release. Under these conditions, the only way the transferase could make a large polymer was to never release it. The enzyme still formed long polymers, suggesting that GlfT2 used a processive process. Yet how could an enzyme using a processive process make a polymer with specific numbers of monomers?
The enzyme adds a monomeric Gal f that is activated for transfer by the addition of a small molecule, UDP (similar to ADP). The Gal f is transferred to the growing glycan which has a very long lipids attached to one end as part of its normal synthetic pathway. They attached different length lipids to the first monomer. They found that if the lipid was too small the enzyme would use a distributive process and the longer the lipid the longer the carbohydrate. This indicates that there is an allosteric lipid binding site on the enzyme, which helps hold the polymer while the enzyme works on it. The lipid, effectively a substrate tether, would effectively decrease the apparent Kd (and perhaps Km) of the growing glycan for the enzyme. The carbohydrate will release from the enzyme and stop growing when the rate of dissociation increases beyond the rate of elongation. This means that the stronger the binding between the enzyme and the lipid the longer the carbohydrate polymer. The way to increase the strength of the lipid enzyme interaction is to increase the length the lipid so the length of the polymer is proportional to the length of the lipid. Hence the length of the lipid that acts as an anchoring for the growing polymer is what determines the length of the carbohydrate, and explains how there can be control in carbohydrate polymerization without the need of a template.
D. General Links and References
![]() Functional Glycomics Gateway (Nature)
Functional Glycomics Gateway (Nature)
![]() Glycan Binding Protein database - CFG (similar to Swiss-Prot for
proteins and GenBank for genes
Glycan Binding Protein database - CFG (similar to Swiss-Prot for
proteins and GenBank for genes
![]() Kyoto Encyclopedia of Genes and Genomes Glycan Database
Kyoto Encyclopedia of Genes and Genomes Glycan Database
D. General Links and References
May, J. et al. A tethering mechanism for length control in a processive carbohydrate polymerization. PNAS 106, 11851(2009)
Hamilton, S. et al. Production of Complex Human Glycoproteins in Yeast. Science, 301, pg 1244 (2003)
Bertozzi., C. and Kiessling , L. Chemical Glycobiology (review) Science 291, pg 2397 (2001)
Helenius and Aebi. Intracellular Functions of N-Linked Glycans. Science. 291, pg 2364 (2001)