Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
D: PROTEIN FOLDING AND STABILITY
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 3/1/16
|
Learning Goals/Objectives for Chapter 2D: After class and this reading, students will be able to
|
D3. Folding of Single Protein Molecules
Protein folding/unfolding reactions can now be studied on single protein molecule using an optical tweezer, as illustrated in the figure below based on a study of RNase H from E. Coli (Cecconi et al, 2005). RNaseH was mutated to contain two Cys residues at positions 4 and 155 (near the ends of the chain). This allowed the protein to be covalently linked through the free sulfhydryl of Cys to two dsDNA molecules (500 bp) chemically modified at one end to form covalent bond to the Cys- containing protein. The CD spectra of the protein linked to the DNA tether was identical to the unmodified protein. The DNA linkers were linked to two different styrene beads though different chemistries to produce the structure shown below.
Figure: Unfolding of RNase H with Optical Tweezers
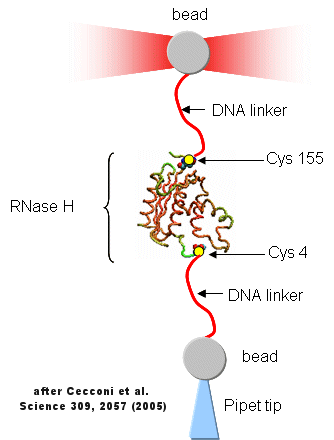
The enzyme was active as shown by enzyme activity measurements. The protein can be stretched and contracted by moving the bead connected to the pipet relative to the bead in the optical trap. As the force (measured in piconewtons) is increased, the molecule is stretched. As a control, the two beads were connected directly with DNA handles without the protein. In the control case, a graph showing extension between the beads vs force increases slowly in a curvilinear fashion at first and then linearly. When the protein is inserted, differences are observed. Upon stretching and relaxing two transitions (shifts) were observed with RNase H that are not seen with DNA alone. These occurred at about 19 pN (an abrupt shift) and a smaller one at 5 pN. These transitions are consistent with the N→D and an I --> D transition , as shown in the figure below (adapted from Cecconi et al). These data are consistent with previous studies of this protein which show the the central, stable core of the protein folds first.
Figure: RNaseH Folding Transitions: Optical Trap
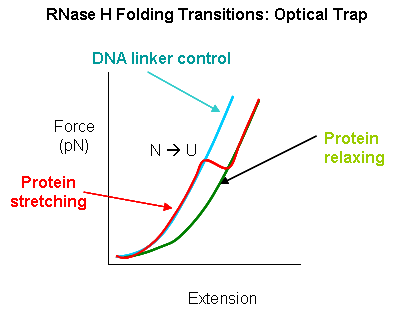
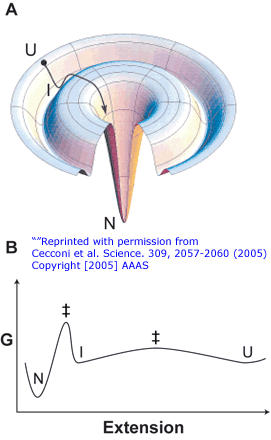
Figure: Folding Lanscape of RNase H: Optical Tweezers
Navigation
Return to Chapter 2D: Protein Folding and Stability
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2D: Protein Folding and Stability

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.