Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
D: PROTEIN FOLDING AND STABILITY
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 3/1/16
|
Learning Goals/Objectives for Chapter 2D: After class and this reading, students will be able to
|
D8. Protein Transport Across Membranes
How does a protein "decide" its final location after synthesis? Protein synthesis occurs in the cytoplasm, but proteins may end up outside of the cell, in cell membranes, internalized into various organelles, or remain in the cytoplasm. How is the decision made? There must be signals in the protein which target proteins to various sites in a cell, where processing can occur. Proteins that are destined for secretion or plasma membrane insertion typically have a signal peptide at the N-terminus which binds to a signal recognition particle in a cotranslational process which temporarily arrests translation. This complex docks to signal recognition complex docking sites in the endoplasmic reticulum membrane, where translation continues as the nascent polypeptide extends through a protein pore in the ER membrane. Gunter Blobel won the Noble Prize in Medicine in 1999 for "for the discovery that proteins have intrinsic signals that govern their transport and localization in the cell".
Figure: Overview - Synthesis of Protein Destined for Secretion
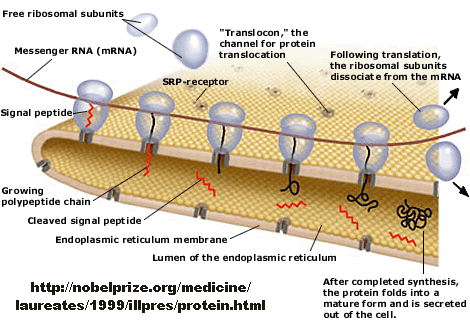
Figure: Signal Recognition Particle Complex
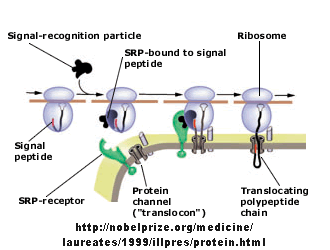
Figure: Sec Dependent Pathway for Post-translational translocation
(reprinted with permission from Kanehisa Laboratories and the KEGG project: www.kegg.org )
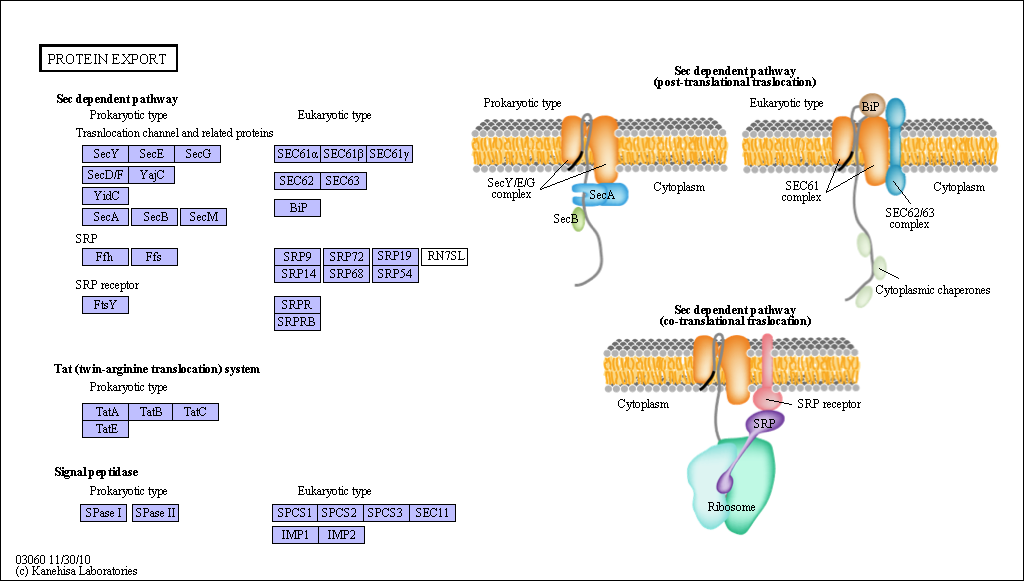
If destined for secretion, it enters the lumen of the ER. Proteins destined for insertion into the cell surface membrane gets "stuck" in the ER membrane, and through a process of vesiculation merges with the Golgi and eventually with the cell surface membrane. Proteins that are taken into organelles like mitochondria are done so in a post-translational process that requires facilitation by protein chaperones. Final protein folding occurs inside the organelle. In both cases, nonnative proteins pass through the membrane after which final folding occurs.
An intriguing question is how the decision is made to keep a protein either in the membrane or allow it to pass through completely (in the case of proteins destined for secretion). Hessa et al investigated this "decision-making" process by studying the eukaryotic membrane pore protein complex, Sec 61 translocon (show in the above figures), whose activity must be closely regulated with the folding of the growing protein. In studying this process, they considered three local regions in a membrane: the hydrophobic region comprised of the nonpolar acyl tails of membrane lipids, the interfacial region in the vicinity of the polar head groups, and the aqueous regions (bulk water) on each side of the head groups. A 19 amino acid peptide was used as the experimental model protein which was added to the translocon. This size was chosen since it is just long enough to span the hydrophobic part of the membrane if the peptide were in an alpha-helical conformation (which is common in membrane-spanning proteins). They varied the proportion of amino acids that tend to partition into each of three regions and studied the disposition of the peptide after interaction with membrane and translocon. To test if the results were consistent with the thermodynamics of amino acid partitioning into nonpolar environments (and not kinetic considerations), they used the Wimley and White hydrophobicity scale, based on the free energy of transfer of amino acid side chains into nonpolar environments, to predict target peptide disposition with the membrane. The table below shows the propensity of amino acids to be in each region at equilibrium, based on this hydrophobicity scale.
Table: Amino Acid Partitioning Into Membrane Regions
| Region | Amino Acids |
| Bulk water | Arg, Asn, Asp, Gln, Glu, His, Lys, Pro |
| Bulk water + interfacial | Ala, Cys, Gly, Ser, Thr |
| Interfacial | Tyr |
| Hydrophobic | Ile, Leu, Met, Phe, Trp, Val |
Their experimental result were in concordance with those predicted using
the above scale. If a polyalanine 19 mer was used, no insertion was
observed. With five leucines in the peptide, almost 90% inserted into
the membrane. The results would be modeling using a two-state
equilibrium:
Peptide inserted <==> Peptide translocated.
They
then substituted each of the twenty amino acids into a given position into a
target peptide and used the results to develop an empirical scale for
membrane transfer, not one based on simple transfer to nonpolar medium.
This new scale matched the hydophobicity scale, suggesting insertion and
transfer decisions where based on thermodynamics of side chain partitioning.
They also varied the position of the varied amino acid in the test peptide.
If the amino acid favored the bulk and/or interfacial region, the peptide
would be inserted if that amino acid were at the end of the peptide, not the
middle. For translocation, the peptide had to be amphiphilic with one face
polar and the other nonpolar.
They developed a simple equilibrium model to show the processes involved, as shown below in a top-down view of the membrane.
Figure: Translocon Equilibrium Model
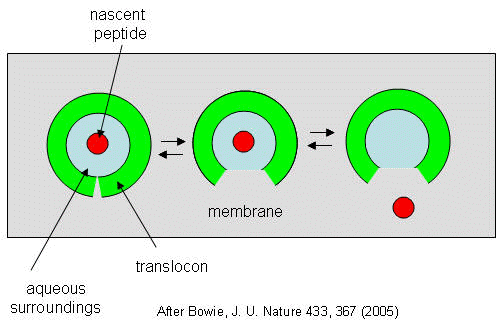
The translocon, shown in green, has a water-filled pore but also a sidewise opening toward the membrane interior. The target peptide enters the pore. Transient conformational changes in the pore expose the peptide to the nonpolar membrane core. The target peptide samples both the aqueous and nonpolar environments and partitions into them based on considerations mentioned above. If it partitions more favorably into the hydrophobic core, it will do so and cause the peptide to become membrane bound. Otherwise it will pass through to the other side. This can be modeled as an equilibrium process if the rate of translocation is slow compared to the rates of translocon conformational change and environmental sampling by the peptide. Obviously, the process becomes more complicated if the target is a large protein.
![]() Jmol:
Updated SecYEB Protein Translocase
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated SecYEB Protein Translocase
Jmol14 (Java) |
JSMol (HTML5)
Bacterial toxin proteins also have evolved ways to pass through a cell membrane, again in a nonnative state, through a protein channel in the membrane. Krantz et al have recently worked out details of how the anthrax toxin protein moves through eukaryotic cell membranes. Three anthrax proteins are involved. One is a "prepore" protein that binds to specific proteins on the cell membrane, where it is activated by limited proteolysis to form a pore protein which assembles into the homoheptamer prepore in the membrane. Two other proteins secreted by the bacteria, lethal factor and edema factor, bind to the heptamar complex and the whole assembly is then taken up into the cell by invagination to form a vesicle with the pore complex in the membrane. This vesicles fuses with a lysosome in the cell, and upon acidification, a conformational change occurs in the prepore complex to activate it. The lethal and edema factors unfold partially, possibly to a molten globule state, and are then passed though the pore into the cell where they exert their toxic influences. An electrochemical potential gradient (which we will discuss later in the semester) is required for passage of the factors through the membrane. The active pore further unravels the factor protein, facilitating transport.
Krantz et al. studied the pore protein by mutating two amino acids, Phe427 and Ser 429, on each monomer of the pore to Cys. They then postranslationally modified the Cys with [2-(trimethylammonium) ethylmethanethiosulfonate and observed effects on ion conductance of the pore and pore conformations. They noted that when both residues were mutated and chemically modified, that ion conductance was blocked, suggesting that these side chains were localized in the narrowest part of the channel. When Phe 427 alone was mutated to smaller side chains (Ala), ion conductance increased but transfer of peptides from the factor proteins was inhibited. This suggested that an aromatic ring in the narrow part of the channel opening participates in translocation of bacterial proteins through the membrane. They then analyzed the transport of a variety of small molecules with varying hydrophobicity through the wild type pore. Their results were consistent with the binding of the molecules through hydrophobic and aromatic electron interactions. They suggest a mechanism of transport consistent with their data in which the unfolded protein "ratchets" through the pore, which promotes factor protein unfolding to expose more hydrophobic groups to the nonpolar aromatic ring in the pore. This mechanism is similar to how the chaperone complex GroEL/GroES unfolds protein in its large central cavity, in a process which requires the chemical potential released by hydrolysis of ATP, not a transmembrane potential. In addition, the Sec61 translocon in the inner membrane of bacteria and in eukaryotic ER membranes also has a pore containing a ring of hydrophobic groups (Ile).
Navigation
Return to Chapter 2D: Protein Folding and Stability
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2D: Protein Folding and Stability

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.