Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
F: THERMODYNAMICS AND IMFs IN PROTEIN STABILITY
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 3/2/16
|
Learning Goals/Objectives for Chapter 2F: After class and this reading, students will be able to ...
|
"You could not step twice into the same river" - Heraclitus of Ephesus (c.535 BC - 475 BC)
F1. Introduction to Protein Stability
This material is not easy, and is perhaps the most intellectually challenging of the entire book. Most of this review comes from an article by Ken Dill, Biochemistry, 29, 7133-7155 (1990). A more recent reanalysis that comes to a significantly different conclusion with respect to the role of H bonds in protein folding and stability, written by Pace, Biochemistry. 40, pg 310 (2001), is discussed at the end.
To summarize, it now appears that both the hydrophobic effect and H bonds appear to drive protein folding and promote protein stability. Extrapolating from the results of studies of the transfer of small model H bond donors/acceptors and hydrophobic molecules from water to nonpolar solvents, it would appears that H bond interactions (as well as ion ..ion interactions) do not drive protein folding per se. Rather, the biggest contributors to stabilization of the native state are the hydrophobic effect and the van der Waals interactions among the tightly packed buried atoms of the protein. However, from recent studies (Pace) of mutant proteins made through site-specific mutagenesis, it appears that H bonds contribute significantly to protein folding and stability, and may make a greater contribution to stability of the native state than the hydrophobic effect. The main factor which opposes folding is chain conformational entropy. These positive and negative factors sum up to a small negative DG favoring protein folding, implying marginal stability of the native protein at normal temperatures.
What types of intermolecular forces might act within a protein and between proteins and solvent molecules that would cause a protein to fold spontaneously to a unique 3D structure? These forces can be long range (ion-ion, ion dipole, or dipole-dipole) or short range (van der Waals repulsive and attractive forces). The interactions can be local (between adjacent amino acids in the linear sequence) or nonlocal (between sequences separated in the linear sequence but brought close together in 3D space). Clues as to what stabilizes the tertiary structure of a native protein can be gained by subjecting proteins to agents that unfold or denature a protein. Such agents include extremes of pH, high concentrations of some salt solutions or organic solvents, and temperature extremes. Such experiments show that native proteins are only marginally stable (about 0.4 kJ/mol amino acid - or around - 10 kcal/mol for a protein of molecular weight of 10,000 - about 100 amino acids). We will consider the different types of intermolecular forces (ion-ion, H bonds, van der Waals, and the hydrophobic effect) individually and ask if each is a significant driving force for protein folding.
Figure: Diagram showing relative contributions to the DG for protein folding.
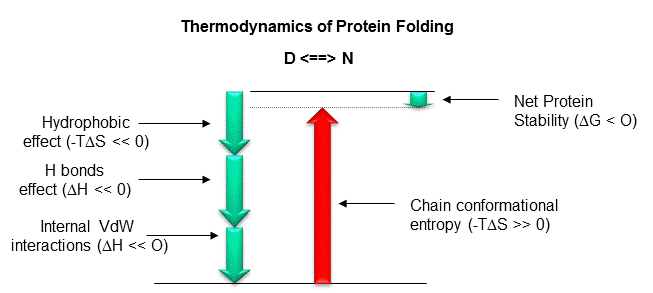
Most of this chapter will deal with H bonding and the hydrophobic effect. A theme of any biochemistry course is that if you can understand the interactions among small molecules, you can apply that knowledge to the understanding of larger molecules like proteins. To understand if H bonds within proteins, often buried in the more hydrophobic interior of the protein, drive protein folding, we will first examine the thermodynamics of H bond formation of a small molecule, N-methylacetamide, in water and in a nonpolar solvent. To understand if the hydrophobic effect, mediated by burying of nonpolar side chains within the more nonpolar center of the protein, drives protein folding, we will examine the thermodynamics of benzene solubility in water. Most recent studies involve the creation of specific mutants at amino acid position that might reveal the contributions of H bonding and the hydrophobic effect to folding and protein stability.
Navigation
Return to Chapter 2F: Thermodynamics and IMFs of Protein Stability
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2F: Thermodynamics and IMFs of Protein Stability

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.