Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
F: THERMODYNAMICS AND IMFs IN PROTEIN STABILITY
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 3/2/16
|
Learning Goals/Objectives for Chapter 2F: After class and this reading, students will be able to ...
|
F2. Ion - Ion Interactions
These could be investigated by altering pH or ionic strength. Why is that?
a. General Charge Interactions - Proteins denature at extremes of pH. At these extremes, proteins have a maximal positive or negative charge, as evident in graphs which show denaturation temperature vs pH for proteins.
Figure: Denaturation temperature vs pH for proteins
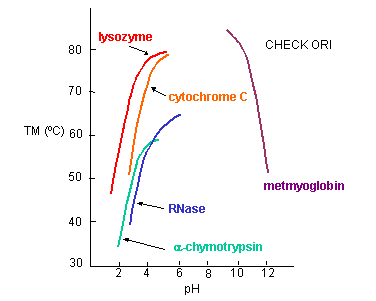
Under extremes of pH (but not so great as to catalyze peptide bond cleavage, electrostatic repulsions would cause the protein to denature. The folded, compact state has an increasing charge density at pH extremes, which could be alleviated by unfolding to a less dense state. But what about specific charge pair interactions? In contrast to the general charge interactions, these might actually stabilize a protein. Are they the predominant factor that determines stability?
b. Specific Charge Interactions (charge pairs) - If ion pairs are the source of protein stability, you would expect that high salt could disrupt them, and lead to denaturation. Although some salts do denature proteins, other stabilize them. Other evidence argues against this idea. Ion pairs are not conserved in evolution. In addition, the number of ion pairs in proteins is small (approx. 5/150 residues, with one of those buried). Also, the stability of a protein shows little dependence on pH or salt concentration (at low concentrations) near the isoelectric point, the pH at which proteins have a net zero charge.
The overall charge state affects not only the stability of a protein but also its solubility. Proteins are most insoluble at their isoelectric point since at that pH (where they have a net 0 charge) the proteins experience the least electrostatic repulsion and are most likely to aggregate and precipitate. Low salt concentration also promotes insolubility. Mutagenesis studies show that solubility can be increased by replacing nonpolar groups on the surface with polar ones. Pace (2009) cites studies on RNase Sa which has a maximally exposed Thr 76. If it is replaced with Asp, the solubility increased to 43 mg/ml but if it is replaced with Trp, it decreased to 3.6 mg/ml. His, Asn, Thr and Gln have a negative effect on solubility near the pI compared to Ala, a surprising result. Similar results were obtained compared to Ala when Arg and Lys were used. Smaller side chains, Asp and Ser, at position 76 increased the solubility over Ala.
Navigation
Return to Chapter 2F: Thermodynamics and IMFs of Protein Stability
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2F: Thermodynamics and IMFs of Protein Stability

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.