Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
G: PREDICTING PROTEIN PROPERTIES FROM SEQUENCES
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 3/9/16
|
Learning Goals/Objectives for Chapter 2G: After class and this reading, students will be able to:
|
G4. Prediction of Membrane Protein Structure
So far we have discussed predominantly globular proteins that are soluble in water. Proteins are also found associated with membranes. Two major classes of membrane proteins are found in nature.
- peripheral membrane proteins: water soluble proteins bound reversibly and non-covalently to the membrane through electrostatic attractions between charged polar head groups of the phospholipids and the protein. These proteins can often be released from the membrane by addition of high salt, since they are often attracted to the bilayer by electrostatic interactions between charged phospholipid head groups and polar/charged groups on the protein surface.
- integral membrane proteins: actually insert into the bilayer. These can be released from the membrane and effectively solubilized by the addition of single chain amphiphiles (detergents) which form a mixed micelle with the integral membrane protein. Nonionic detergents (Trition X-100, octylglucoside, etc) are often used in the purification of membrane proteins. Ionic detergents (like SDS) not only solubilize the integral membrane proteins, but also denature them.
Figure: Types of membrane proteins
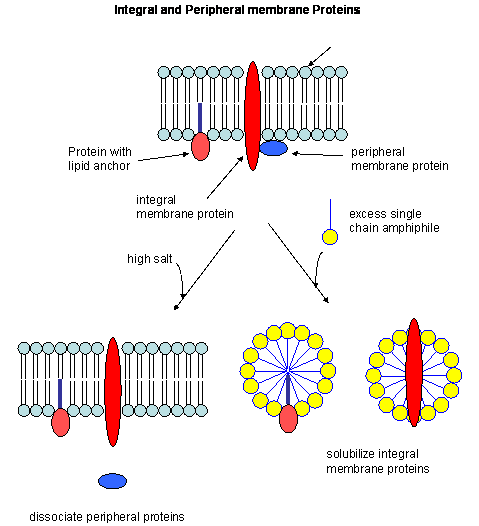
In some of these integral membrane proteins, large extracellullar and intracellular domains of the protein are present, connected by the intramembrane regions. The intramembrane spanning region often consists of either a single alpha helix, or 7 different helical regions which zig-zag through the membrane. These transmembrane sequences can readily be determined through hydropathy calculations. For example, consider the integral membrane bovine protein rhodopsin. Its 348 amino acid sequence (in single letter code) is shown below:
MNGTEGPNFYVPFSNKTGVVRSPFEAPQYYLAEPWQFSMLAAYMFLLIMLGFPINFLTLY
VTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG
GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIP
EGMQCSCGIDYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTVKEAAAQQQES
ATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAV
YNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTETSQVAPA
Rhodopsin hydropathy plot calculations shows that is contains seven transmembrane helices which wind through the membrane in a serpentine fashion..
Figure: Rhodopsin hydropathy plot
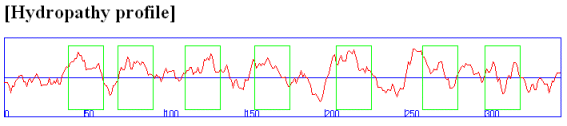
Figure: seven transmembrane helices
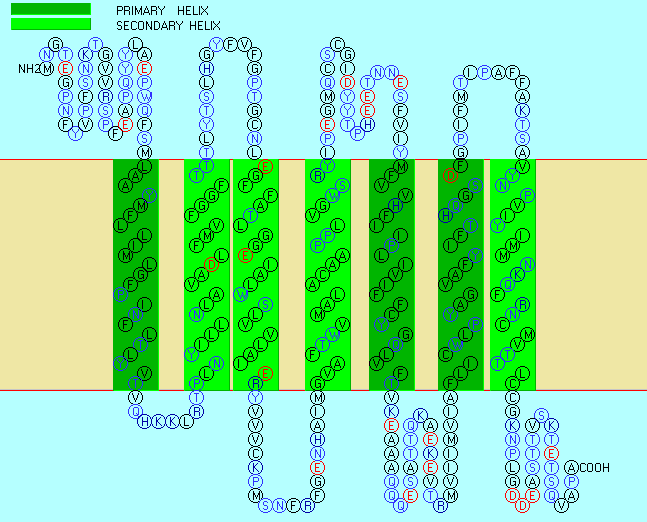
Rhodopsin Hydropathy Results
| No. | N terminal | transmembrane region | C terminal | type | length |
| 1 | 40 | LAAYMFLLIMLGFPINFLTLYVT | 62 | PRIMARY | 23 |
| 2 | 71 | PLNYILLNLAVADLFMVFGGFTT | 93 | SECONDARY | 23 |
| 3 | 113 | EGFFATLGGEIALWSLVVLAIER | 135 | SECONDARY | 23 |
| 4 | 156 | GVAFTWVMALACAAPPLVGWSRY | 178 | SECONDARY | 23 |
| 5 | 207 | MFVVHFIIPLIVIFFCYGQLVFT | 229 | PRIMARY | 23 |
| 6 | 261 | FLICWLPYAGVAFYIFTHQGSDF | 283 | PRIMARY | 23 |
| 7 | 300 | VYNPVIYIMMNKQFRNCMVTTLC | 322 | SECONDARY | 23 |
In summary, hydropathy plots are hence useful in finding buried regions in water soluble proteins, transmembrane helices in integral membrane proteins as well as short stretches of polar/charged amino acids that might form surface loops recognizable by immune system antibodies. The window size used in hydropathy plots would obviously affect the calculated results. Windows of 20 amino acids are useful to determine transmembrane helices while windows of 5-7 amino acids are used to find surface-exposed hydrophilic sites.
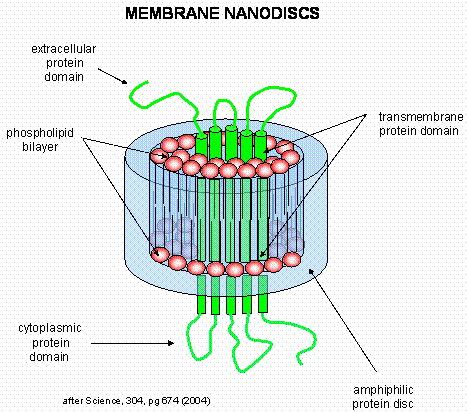
Membrane proteins call be solubilized by addition of single chain amphiphiles (detergents). The nonpolar tails of the detergents interact with the hydrophobic transmembrane domain of the membrane protein forming a "mixed" micelle-like structure. Nonionic detergents like Triton X-100 and octyl-glucoside are often used to solubilize membrane proteins in their near native state. In contrast, ionic detergents like sodium dedecyl sulfate (with a negatively charged head group) denature proteins during the solubilization process. To study membrane proteins in a more native-like environment, proteins solubilized by nonionic detergent can be reconstituted into bilayer liposome structures using methods similar to those from Lab 1 in which you prepared dye-capsulated large unilamellar vesicles (LUVs). However, it can be difficult to study the intra- and extracellular domains of membrane proteins in liposomes, given that one of those domains is hidden inside the liposome. A novel technique that removes this barrier was recently developed by Sligar. He created an amphiphilic protein disc with an opening in the center. The inner opening is lined with nonpolar residues, while the outer surface of the disc is polar. When the discs were added to phosphlipids, small bilayers formed inside the disc. Membrane proteins like the b-2 adrenergic receptor could be reconstituted in the nanodisc bilayers, allowing solvent exposure of both the intracellular and extracellular domains of the receptor protein.
Figure: Nanodisc with membrane protein
- Experimentally Determined Hydropathy Scales
- Protein Sequence Structural Features
- Membrane Protein Resources
- Membrane Proteins of Known 3D Structure
- 57 Different Amino Acid Scale Predictors from ExPASy
Navigation
Return to Chapter 2G: Predicting Protein Properties from Sequences
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2G: Predicting Protein Property from Sequences

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.