Cut Site Selection by the Two Nuclease Domains of the Cas9 RNA-guided Endonuclease. Hongfan Chen, Jihoon Choi and Scott Bailey.The Journal of Biological Chemistry, 289, 13284-13294 (2014)
Questions from article
1. Investigators studied the type II CRISPR-Cas system of S. thermophilus LMG18311.
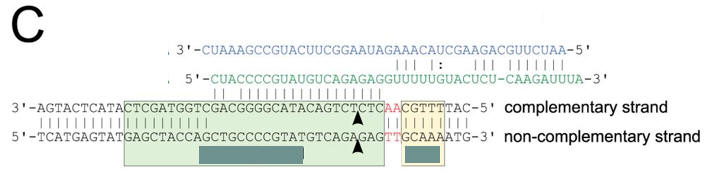
a. Figure A shows a Coomassie Blue-stained SDS-polyacrylamide gel of Cas9, Cas9 D9A, Cas9 D599A, and Cas9 D9A,D599A where D is aspartic acid and A is alanine and D#A reflects the site of the specific mutation where D was changed to A. Why did they perform this experiment?

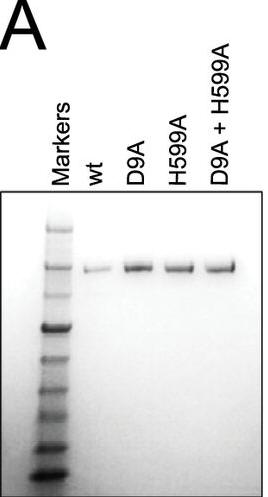
b. To confirm the PAM sequence for LMG18311 Cas9, the authors performed BLAST searches to identify potential protospacers in viral and plasmid genomes that matched any of the 33 spacer sequences from CRISPR-1. This search generated 41 unique target sequences, from the genomes of bacteriophage known to infect S. thermophilus. We then aligned 50-nucleotide segments from the identified target genomes, inclusive of the 30-nucleotide protospacer and 10-nucleotide flanking regions. Figure b shows B, logo plot revealing the PAM for LMG18311 Cas9. The positions of the protospacer, PAM, and linker are indicated.
What is the most likely consensus sequence of the PAM sequence? How far is it away from the protospacer:
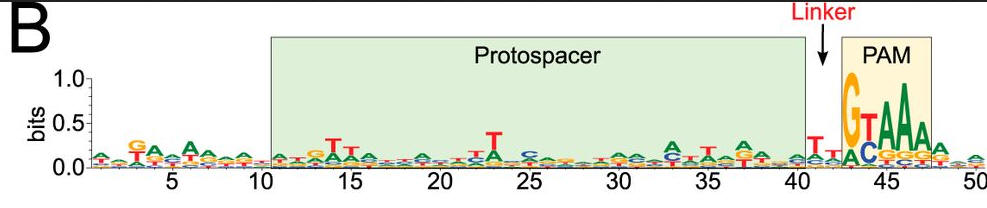
c. Figure C shows a schematic representation of the CRISPR nucleic acid complexes. Label the target DNA, crRNA, tracrRNA,, protospacer, PAM, and linker. Draw in a connecting piece of RNA if you wish to create a single guide RNA containing both the crRNA and tracRNA. What is the specific PAM sequence in this figure.