Protein Symmetry
Java version
HTML 5 version (does not require Java; downloads and moves
slowly)
I. Introduction
Proteins of the C2, C4, D2, and D4 symmetry
point groups are below with operations of the characteristic
symmetry elements of the point groups. The subunits of the
proteins are color coded so the change in orientation is clearly
observed.
cyclic (Cn) - contain one single Cn
rotation axis. In this point group note that the
n in Cn is equal to the number of monomers and the angle of
rotation is 360o/n..
dihedral (Dn) - These have mutually
perpendicular rotation axes. Specifically they contain at least 1 C2
axis perpendicular to a Cn axis (Canter and Schimmel.
Biophysical Chemistry - Part 1). The minimal number of
subunits is n. Most protein oligomers fall into this
category.
Click
on the symmetry element button below to see each individual
protein undergo the symmetry operation and compare the rotated
protein on the left to the fixed orientation on the right.
Note: The Jmol command used to carry
out the rotations also rotate the axes. We are working to
fix this problem so the superpositions can be better visualized.
For more information see
Biochemistry Online: Chapter 5C -
Model Binding Systems
II. General Structure
C2 Symmetry: Alcohol Dehydrogenase
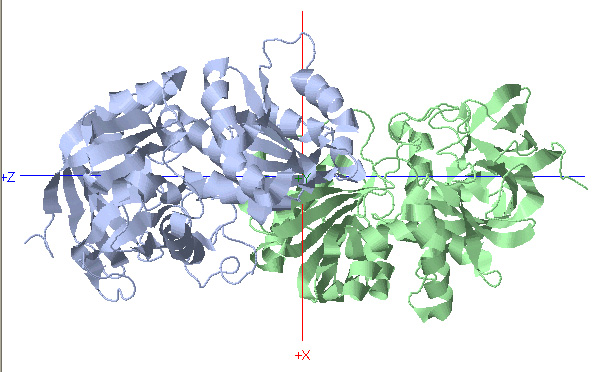
Rotational Axis
*Rotate protein 180 degrees about Y-axis
C4 Symmetry: Neuraminidase
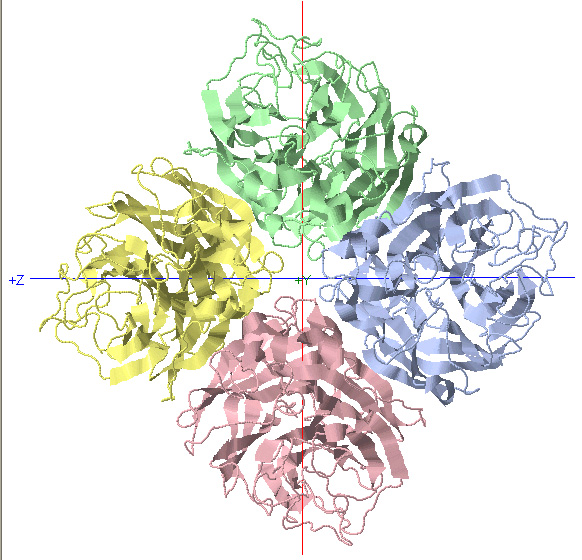
C4 Rotational Axis
*Rotate protein 90 degrees about Y-axis
D2 Symmetry: Phosphofructokinase

C2 Rotational Axis
*Rotate protein 180 degrees about Y-axis
D4 Symmetry: Glycolate Oxidase
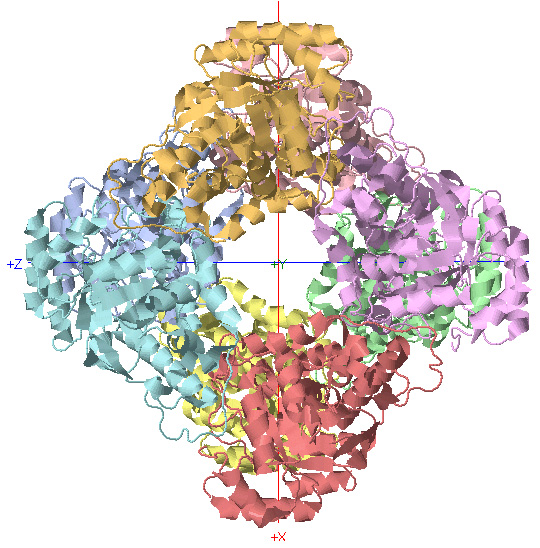
C4 Rotational Axis
*Rotate protein 90 degrees about Y-axis
Horizontal Plane
*Rotate protein 90 degrees about Z-axis to see mirror
image subunits above and below Z-axis (plane is perpendicular to Y-axis)