08/14/2014
French translation by Kate Bondareva
Central Dogma of Biology: DNA --> RNA --> Protein
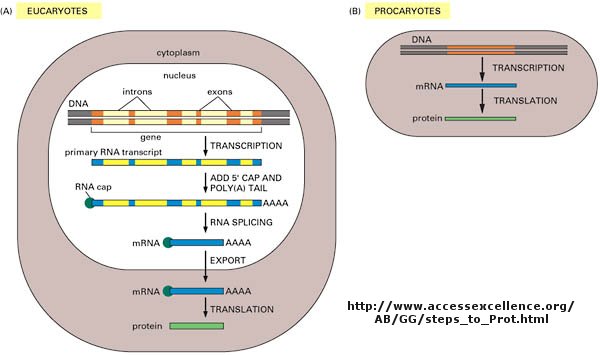
DNA is the carrier of genetic information in organisms. What does that mean? Large molecules in organism can have many functions: they can provide structure, act as catalyst for chemical reactions, serve to sense changes in their environment (leading to immune responses to foreign invaders and to neural responses to stimuli such as light, heat, sound, touch, etc) and provide motility. DNA really does none of these things. Rather you can view it as an information storage system. The information must be decode to allow the construction of other large molecules. The other molecules are usually proteins, another class of large polymers in the body. Chromosomes are located in the nucleus of a cell.
Replication
Chromosomes are located in the nucleus of a cell. DNA must be duplicated in a process called replication before a cell divides. The replication of DNA allows each daughter cell to contain a full complement of chromosomes.
Transcription
The actual information in the DNA of chromosomes is decoded in a process called transcription through the formation of another nucleic acid, ribonucleic acid or RNA. The RNA, made by the enzyme RNA polymerase, is complementary to one strand of the DNA. RNA differs from DNA in that RNA contains a ribose, not deoxyribose, sugar in its backbone. In addition, RNA lacks the base T. It is replaced, instead, with the base U, which is complementary to A (as T is complementary to A in DNA). The RNA formed acts as a messenger, which passes from the nucleus into the cytoplasm of the cell. In fact, this type of RNA is often called messenger RNA, mRNA. Since the information in a nucleic acid (DNA) is converted into information in the form of another nucleic acid (RNA), this process is called transcription (since the language is still the same, such as when you transcribe a written speech in English into written English).
Translation
The information from the DNA, now in the form of a linear RNA sequence, is decoded in a process called translation, to form a protein, another biological polymer. The monomer in a protein is called an amino acid, a completely different kind of molecule than a nucleotide. There are twenty different naturally occurring amino acids that differ in one of the 4 groups connected to the central carbon. In an amino acid, the central (alpha) carbon has an amine group (RNH2), a carboxylic acid group (RCO2H), and H, and an R group attached to it. Since the information in a nucleic acid (RNA) is converted into information in the form of a different molecule, a protein, this process is called translation (since the language of nucleic acids is changed to that of proteins, such as when you translate English into Chinese).
In contrast to the complementarity of DNA and RNA (1 base in RNA complementary to 1 base in DNA), there is not a 1:1 correspondence between a base (part of the monomeric unit of RNA) in RNA to the monomer in a protein. After much work it was discovered that a contiguous linear sequence of 3 nucleotides in RNA is decoded by the molecular machinery of the cytoplasm with the result that 1 amino acid is added to the growing protein. Hence a triplet of nucleotides in DNA and RNA have the information for 1 amino acid in a protein. That there was not a 1:1 correspondence between nucleotides in nucleic acids and amino acids in proteins was evident long ago since there are only 4 different DNA monomers (with A, T, G, and C) and 4 different RNA monomers (with A, U, G, and C) but there are 20 different amino acid monomers that compose proteins.
Now, it turns out that not all the information in the DNA sequence of a organism encodes for a protein. In fact only about 2% of the 3 billion base pairs seem to be transcribed into RNA which can be translated into protein. The function of the rest of the DNA is at present uncertain. How does the molecular machinery of the cell know which part of the DNA encodes for proteins. It turns out that there are unique DNA sequences at the beginning and end of the part of the DNA sequence that codes for a protein. Proceed down the DNA of a chromosome and suddenly you come to those signals, which are recognized by the cells machinery. A complementary RNA is made from that section, and the complementary RNA is then decoded into a single protein. Continue further down the DNA sequence and another such coding sequence is found, which can be transcribed into a mRNA, which then can be translated into another unique protein. In all there are about 30,000 such sections of DNA in all the chromosomes that encode the information for 30,000 unique proteins. These unique coding sections of DNA that ultimately are transcribed into unique mRNA which are translated into unique proteins are called genes. For our purposes, we conclude that one gene has the information for one protein. Each of the protein differ from each other in both length, and the specific sequence of amino acids in the protein. The DNA is indeed the blueprint of the cell. What determines the actual characteristics of the cells are the actual proteins that are made by the cell.

Not only must DNA be transcribed into DNA, but the genetic information in the DNA must be replicated before a given cell divides, so that the daughter cells both contain the same genetic information. In replication, the dsDNA separate, and an enzyme, DNA polymerase, makes complimentary copies of each strand. The two resulting dsDNA strands separate to different daughter cells during division. The process where by DNA is replicated when cells divide, and is transcribed into RNA which is translated into protein is called the Central Dogma of Biology. (disregard tRNA, rRNA, and snRNA in the preceding web link)
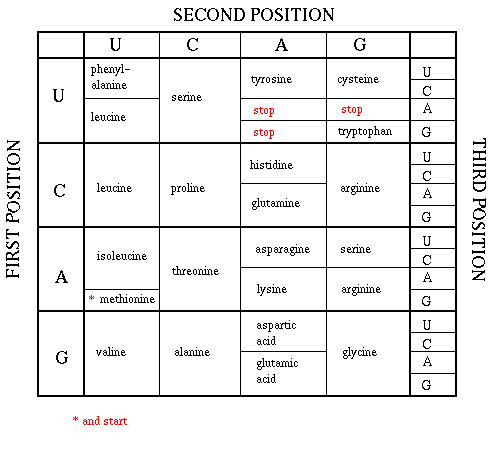
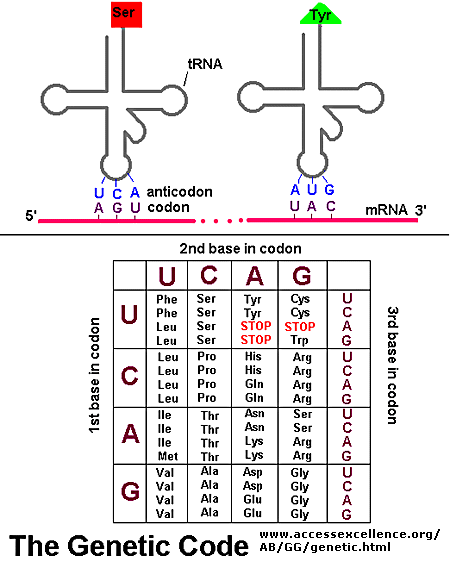
As mentioned above, each amino acid is specified by a particular combination of three nucleotides in RNA. The three bases are called a codon. The Genetic Code consists of a chart which shows what triplet RNA sequence or codon in mRNA codes for which of the 20 amino acids. One of the codon codes for no amino acids and serves to stop the synthesis of the protein from the mRNA sequence. The genetic code is shown below:
GENETIC CODE
Determining the protein sequence from a DNA sequence.
For a given gene, only one strand of the DNA serves as the template for transcription. An example is shown below. The bottom (blue) strand in this example is the template strand, which is also called the minus (-) strand, or the sense strand. It is this strand that serves as a template for the mRNA synthesis. The enzyme RNA polymerase synthesizes an mRNA in the 5' to 3' direction complementary to this template strand. The opposite DNA strand (red) is called the coding strand, the nontemplate strand, the plus (+) strand, or the antisense strand.
The easiest way to find the corresponding mRNA sequence (shown in green below) is to read the coding, nontemplate, plus (+), or antisense strand directly in the 5' to 3' direction substituting U for T. Find the triplet in the coding strand, change any T's to U's, and read from the Genetic Code the corresponding amino acid that would be incorporated into the growing protein. (The strands below are separated into triplets for ease of visualization.) An example is show below.
DNA 5' A G G C C T T C G A A C G G G A T G G A A T G A 3' DNA 3' T C C G G A A G C T T G C C C T A C C T T A C T 5'
RNA 5' A G G C C U U C G A A C G G G A U G G A A U G A 3'
PROT N ARG PRO SER ASP GLY MET GLU STOP
END BASIS REVIEW CENTRAL DOGMA OF BIOLOGY
Proteins
In contrast to the linear polymers of DNA and RNA, proteins (linear polymers of amino acids) fold in 3D space to form structures of unique shapes. Each unique protein sequence (of a given length and sequence of amino acids) folds to a unique 3D shape. Hence there are about 30,000 proteins of different shapes in humans. Not only do proteins have unique shapes, but they also have unique nooks and crannies and pockets which allow them to bind other molecules. Binding of other molecules to proteins or DNA initiates or terminates the function of the protein or nucleic, much like an on/off switch. The example below show different protein structures, some of which have small molecules or large molecules (like DNA) bound to them. Some common motifs are found within the 3D structure of the protein. The include alpha helices and beta sheets. These are held together by H-bonds between the slightly positive H on the N in the protein backbone and a slightly negative O further away on the protein backbone. In the Chime models below, use the mouse controls to rotate the molecule. (Also shift L-mouse click will change the size of the molecule). Click on the command in the right hand frame to change the rendering of the proteins. The cartoon view allows a simple way to interpret the overall structure of the main chain.
Mutations
If the DNA sequence in a coding region becomes changed, the resulting mRNA will also be changed, which will lead to changes in the protein sequence. These changes might have no effect and be silent, if the change in the protein does not affect the folding of the protein or its binding to another important molecule. However, if the changes affects either the folding or the binding region, the protein may not be able to perform its usual function. Mutations which substitute nonpolar amino acids for polar/charged ones (or the reverse) have the greatest chance of causing significant changes in structure and/or activity.
If the function was to put on break on cell division, the result might lead the cell to become a cancerous. Likewise, if the normal protein had a role in causing the cell to die after its intended life span, the cell with the mutant protein might not die and more likely become a tumor. The opposite scenarios could happen leading the cell to a premature death.
Check out the model of sickle cell hemoglobin below, which shows how a single base pair change in the DNA can change one amino acid in a protein with lethal consequences.
Genes and Disease
Activation and Repression of Gene Transcription:
Imagine you have been given a one foot string which represents part of a human chromosome. There just happens to be a small dot in the dead center of the string you have been given. It represents the gene for a particular protein called metallothionein. This gene is expressed and the protein metallothionein is made when cells are exposed to toxic heavy metals like mercury. It would be as waste of metabolic energy if all 30,000 genes were expressed (transcribed and translated into proteins) all the time. In the case of dealing with mercury exposure, it would make most sense if the gene for metallothionein, in the absence of mercury, was not expressed or expressed to a small, constituitive level, but in the presence of mercury, its level of expression would increase to deal with the threat to the cell. How might the cell accomplish these two feats:
One of the central questions of modern biology is what controls gene expression. As we have previously described, genes must be "turned on" at the right time, in the right cell. To a first approximation, all the cells in an organism contains the same DNA (with the exception of germ cells and immune cells). Cell type is determined by what genes are expressed at a given time. Likewise, cell can change (differentiate) into different types of cells by altering the expression of genes. The central dogma of biology describes how genes are first transcribed to RNA, and then the mRNA is translated into a corresponding protein sequence. Proteins can then be post-translationally modified, localized to certain locales within the cells, and ultimately degraded. If functional proteins are considered the end-product of gene expression, the control of gene expression could theoretically occur at any of these steps in the process.
Mostly, however, gene expression is controlled at the level of transcription. This makes great biological sense, since it would be less energetically wasteful to induce or inhibit the ultimate expression of a functional protein at a step early in the process. How can gene expression be regulated at the transcriptional level? Many examples have been documented. The main control is typically exerted at the level of RNA polymerase binding just upstream (5') of a a site for transcriptional initiation. Other factors, called transcription factors (which are usually proteins), bind to the same region and promote the binding of RNA polymerase at its binding site, called the promoter. Proteins can also bind to sites on DNA (operator in prokaryotes) and inhibit the assembly of the transcription complex and hence transcription. Regulation of gene transcription then becomes a matter of binding the appropriate transcription factors and RNA polymerase to the appropriate region at the start site for gene transcription. Regulation of gene expression by proteins can be either positive or negative. Regulation in prokaryotes is usually negative while it is positive in eukaryotes.
Most eukaryotic genes have about 5 regulatory sites for binding transcription factors and RNA polymerase. Examples of these transcription factors are show in the figure below.
STRUCTURAL FEATURES OF SPECIFIC DNA BINDING SITES
Since RNA polymerase must interact at the promoter site of all genes, you might expect that all genes would have a similar nucleotide sequence in the promoter region. This is found to be true for both prokaryotic (such as bacteria) and eukaryotic genes. You would expect, however, that all transcription factors would not have identical DNA binding sequences. The sequences of DNA just upstream of the start site of the gene that binds protein (RNA polymerase, transcription factors, etc) are called promoters. The table below shows the common DNA sequence motif called the Pribnow or TATA box found at around -10 base pairs upstream from the start site, and another at -35. Proteins bind to these sites and facilitate binding of RNA polymerase, leading to gene transcripton.
![]() Jmol: TATA
Box Binding Protein
Jmol: TATA
Box Binding Protein
Prokaryotic Promoter Sequences
|
Promoter |
-35 Region |
Spacer | -10 Region | Spacer | RNA start |
| trp operon | TTGACA | N17 | TTAACT | N7 | A |
| tRNAtyr | TTACA | N16 | TATGAT | N7 | A |
| lP2 | TTGACA | N17 | GATACT | N6 | G |
| lac operon | TTTACA | N17 | TATGTT | N6 | A |
| rec A | TTGATA | N16 | TATAAT | N7 | A |
| lex A | TTCCAA | N17 | TATACT | N6 | A |
| T7A3 | TTGACA | N17 | TACGAT | N7 | A |
| consensus | TTGACA | TATAAT |
In addition, in eukaryotes promoters, sequences further upstream called response elements bind specific proteins (such as CREB or cyclic AMP response element binding protein) to further control gene transcription.
Eukaryotic Response Elements (RE)s
|
Regulatory agent |
Module |
Consensus | DNA bound | Factor | Size (daltons) |
| Heat Shock | HSE | CNNGAANNTCCNNG | 27 bp | HSTF | 93,000 |
| Glucocorticoid | GRE | TGGTACAAATGTTCT | 20 bp | Receptor | 94,000 |
| Cadmium | MRE | CGNCCCGGNCNC | . | ? | . |
| Phorbol Ester | TRE | TGACTCA | 22 bp | AP1 | 39,000 |
| Serum | SRE | CCATATTAGG | 20 bp | SRF | 52,000 |
| Antioxidant | ARE | GTGACTCAGC | |||
| Pheromone (fungus) | ACAAAGGGA | ||||
| Hypoxia | HRE |
CCACAGTGCATACGT GGGCTCCAACAGGTC CTCTCCCTCCCATGCA |
Hypoxia Inducible Factor | 826 aa | |
| Peroxisome Proliferator Activated Receptor (PPAR) | PPRE | aGG_CAAAGGT(CG)A | PPAR | 59,000 | |
| Steroid (general) (progesterone, androgen, mineralcorticoids, glucocorticoids | AGAACAxxxACAAGA (inverted repeat) |
Proteins can interact specifically with DNA through electrostatic, H-bond, and hydrophobic interactions. AT and GC base pairs have available H bond donors and acceptors which are exposed in the major and minor grove of the ds DNA helix, allowing specific protein-DNA interactions.
![]() Jmol: Simple
DNA Tutorial (see last selection buttons to see H bond
donors and acceptors in the major grove.
Jmol: Simple
DNA Tutorial (see last selection buttons to see H bond
donors and acceptors in the major grove.
STRUCTURAL FEATURES OF DNA-BINDING PROTEINS
Not any protein can bind specifically to DNA. Analysis of DNA binding proteins shows common motifs are found among them.
PHOSPHORYLATION AND CONTROL OF GENE EXPRESSION
A common way to control gene expression is by controlling the phosphorylation of transcription factors by ATP. This modification might activate or inhibit the transcription factor in turning on gene expression. The added phosphate groups might be necessary for direct binding interactions leading to gene transcription or they might lead to a conformational change in the transcription factor, which could activate or inhibit gene transcription. A recent example of this later case is the control of the activity of the transcription factor p53. p53 has many activities in the cell, a primary one as a suppressor of tumor cell growth. If a cell is subjected to stress that results in genetic damage (an evident which could lead a cell to transform into a tumor cell), this protein becomes an active transcription factor, leading to the expression of many genes, including those involved in programmed cell death and cell cycle regulation. Both of these effects could clearly inhibit cell proliferation. Hence p53 is a tumor suppressor gene. p53 is usually bound to the protein HDM2 which down regulates its activity by leading to its degradation. Stress signals lead to the activation of protein kinases in the cell (such as p38, JNK, and cdc2), causing phosphorylation of serine and threnine amino acids in p53 (Ser 33 and 315 and Thr 81). This leads to the binding of the proteni Pin 1, which changes the shape of p53 and leads to its activation as a transcription factor.