04/17/16
"Ugly giant bags of mostly water" -
The crystal life form describing humans
on Star Trek Next Generation, Home Soil Episode.

04/17/16
"Ugly giant bags of mostly water" -
The crystal life form describing humans
on Star Trek Next Generation, Home Soil Episode.
This book began with the notion that understandings derived from the study of simple molecules can be applied to complex biological macromolecules and systems. We developed an understanding of the structural, thermodynamic, and kinetic properties of the "simplest" biomolecules, including single chain amphiphiles like fatty acids, and double chain ones like phospholipids, and how these properties could explain the propensity of these molecules to form complex lipid aggregates (micelles and bilayers). We extended these ideas to the process of protein folding and the assembly of biological complexity. Is there something intrinsic to the property of molecules such that their localization together in the right microenvironment could lead to a "living cell"? How did life originate? That is the topic of this last capstone chapter.
Defining life is actually quite difficult. Here is a list of requirements that seem reasonable, but other have noted that this list would exclude the mule. Life can self-replicate, self-sustain, evolve, respond to environmental changes, and die. The earliest known fossils (stromatolites from cyanobacteria) are approximately 3.5 billion years old.

We have just finished studying the complex interactions involved in cell signaling. How could they have evolved? Consider the central dogma of biology. In the present biological world, proteins (DNA polymerase, RNA polymerase, transcription factors) are necessary for DNA synthesis, replication and gene transcription. But you need DNA to encode the proteins. This "chicken vs egg" dilemma has been addressed when it was realized that RNA can both carry genetic information as well as enzymatic activity (even at the level of the ribosome used for protein synthesis).

Much work has been done to determine if the building blocks for present biological molecules could have been synthesized early in Earth's history. Amino acids and fatty acids have been found in meteors suggesting the possibility. Earth's early atmosphere would have had little oxygen, so most components should have been reduced. It probably consisted of methane, ammonia, hydrogen and water similar to the atmospheres of other planets in our solar system. The composition of the early atmosphere is still contentious. In 1953 (the same year that Watson and Crick published the structure of double-stranded DNA), Stanley Miller showed that electric discharges (to simulate lightening) in a reducing atmosphere over a "simulated sea" produced many amino acids. Up to 11 different amino acids have been produced in this fashion along with purines and pyrimidines (these required concentrated reaction mixtures) necessary for nucleic acids. Adenine can be produced just through the reaction of hydrogen cyanide and ammonia in an aqueous solution. Other nucleic acid bases can be made with hydrogen cyanide, cyanogen (C2N2) and cyanoacetylene (HC3N).
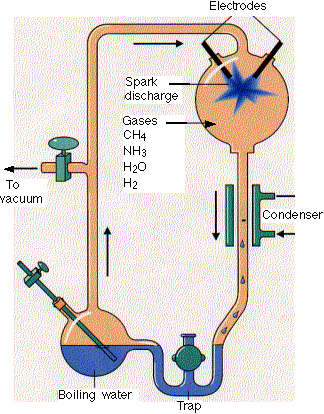
http://www.hencoup.com/Photo%20Stanley%20Miller.jpg
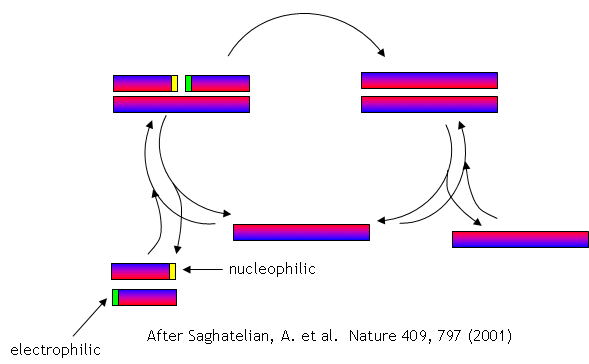
No complex polymers arise through these reactions. However, in 2004, Lehman, Orgel and Ghadiri were able to show that in the presence of carbon disulfide, a gas discharged from volcanoes, homo- and hetero-peptides were produced. Amphiphilic peptides can even catalyze their own formation from peptide fragments, if the fragments are activated. The fragments would bind to the larger "template" peptide through nonpolar actions of the side chains which are oriented along one face of the helical axes. If the fragments bind in a fashion in which the electrophilic end is adjacent to the nucleophilic end of the other peptide fragment, condensation of the two peptide fragments results. The larger template peptide acts as a template (effectively as an "enzyme") in orienting the two fragments for chemical reaction and effectively increasing their local concentration. The reaction of the bound fragments is essentially intramolecular. The reaction even proceeds with amplification of homochirality.
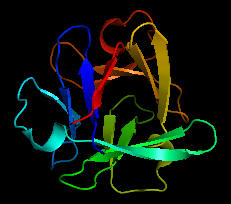
Could the prebiotic amino acids have polymerized into a protein that could fold in a fashion similar to modern proteins? That question has recently been addressed by Longo et al (2013). They asked the question whether the amino acids found in Miller-type prebiotic synthesis mixture and in comets/meteors (Ala, Asp, Glu, Gly, Ile, Leu, Pro, Ser, Thr and Val), a restricted set (10) compared to the present 20 naturally-occurring amino acid, could form a polymer that could fold. Notice that this reduced ensemble of amino acids lacks aromatic and basic amino acids. These proteins would be acidic with a low pI and may have trouble, given the lack of nonpolar aromatic amino acids, in forming a buried hydrophobic core which stabilize proteins. Nevertheless Longo et al were able to synthesize a protein with a slightly expanded set of amino acids (12, including Asn and Gln, with 70% prebiotic amino acids). The structure of one of the proteins, PV2, is shown below. The protein was more stable in 2 M NaCl (compared to 0.1 M) in which it showed a cooperative thermal denaturation with a melting point near 650C using differential scanning calorimetry. The protein had properties similar to those from halophilic organisms that thrive in high salt. These properties include low pIs and high negative charge density, which allows cation-protein interactions in the high salt environment, and lower stability in low salt environments. Earlier oceans were saltier. Halophiles are an example of extremeophiles which are highly represented in archea. Although most halophiles are aerobic, some are anerobic. Perhaps life arose in high salt environments.
Figure: Structure of PV2 protein comprised of a reduced alphabet of mainly prebiotic amino acids.
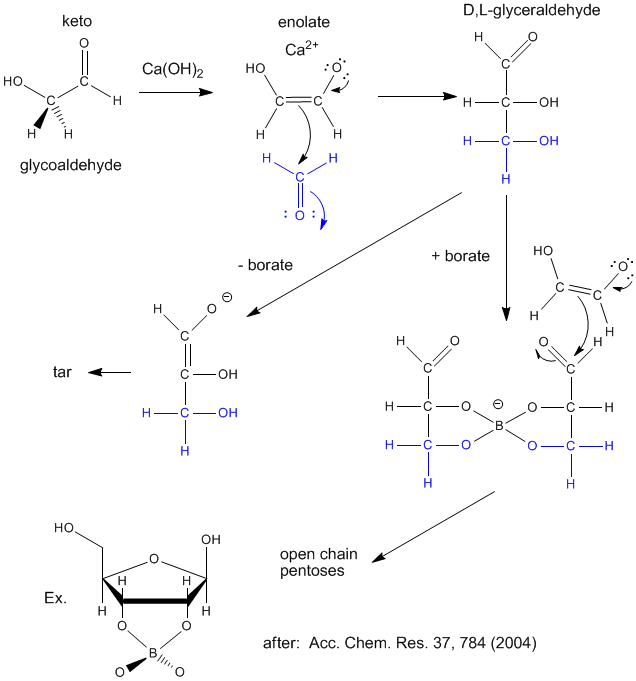
Sugars are required for present energy production but also as a part of the backbone (ribose, deoxyribose) of present genetic material. Many sugars can be synthesized in prebiotic conditions, using carbon based molecules with oxygen, such as glycoaldehyde and formaldehyde (both found in interstellar gases), as shown in the figure below. Glycoaldehyde was recently found in star forming regions of the Milky Way where planets are likely to form. The presence of borate, which stabilizes vicinal OHs on sugars, is required for the production of sugars instead of tars.
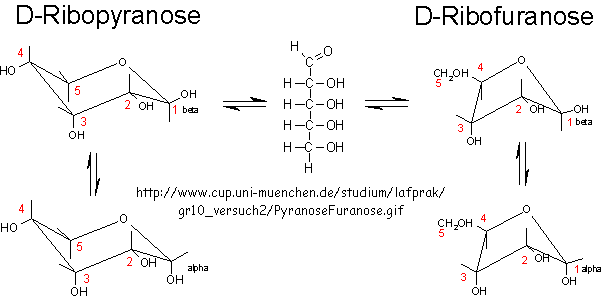
RNA molecules containing sugars such as threose, aldopentopyraonses and hexopyranose can also form stable secondary structures like helices. (Remember, RNA probably preceded DNA as the genetic carrier of information given that is also has enzymatic activity). Is there something special about ribose that made it selected over other sugars for nucleic acids, especially since it is found in low abundancy in the products in synthesis reactions conducted under prebiotic conditions? One probable reason is it unusually high (compared to other sugars) permeability coefficient through vesicles made of phospholipids or single chain fatty acids, as shown below.
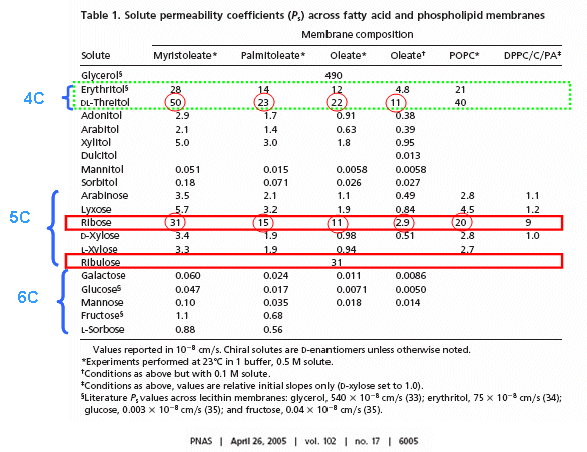
In general, the greater the number of carbon atoms, the smaller the permeability. However, the table above clearly shows large differences in permeability for sugar isomers with the same number of C atoms, and the difference is not affected by the lipid composition. Ribose has markedly elevated permeability compared to other 5C sugars (as do erythritol and threitol among 4 C sugar alcohols). What is so unique about ribose? 20% of the sugar is in the furanose form. Rate constants for ring opening of furanoses are elevated, suggesting greater flexibility. The α-furanose anomer is amphiphilic in that one face is hydrophobic and the other hydrophilic. All of these may promote ribose permeability.
![]()
![]() Jmol:
a-D-ribofuranose
alpha-D-ribopyranose at
http://www.chemeddl.org/resources/models360/models.php
Jmol:
a-D-ribofuranose
alpha-D-ribopyranose at
http://www.chemeddl.org/resources/models360/models.php
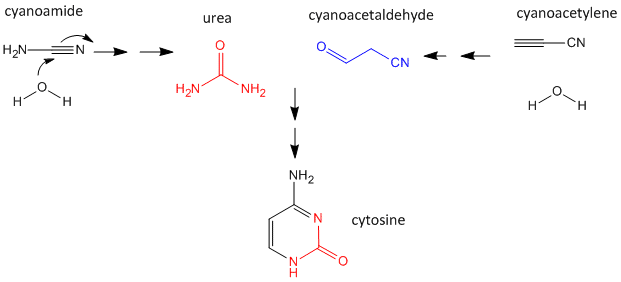
As mentioned above, nucleobases can be made under abiotic conditions with appropriate sources of carbon molecules with nitrogen. These include hydrogen cyanide, cyanoamide (NH2CN) (along with ammonia), hydrogen cyanide, cyanogen (C2N2) and cyanoacetylene (HC3N). Cyanoamide and cyanoacetylene can both react with water to form urea and cyanoacetaldehyde, respectively. The latter two can condense to form cytosine as shown below.
Figure: Abiotic Synthesis of Cytosine
A major problem that has plagued prebiotic researchers is how the product of the carbon-oxygen compound (sugar) links to the product of the carbon-nitrogen compound (nucleobase) to form the nucleoside. Recent research by Powner et al has offered an innovative solution. Instead of forming the sugar and base in separate reaction, and then linking them covalently, the combined molecule could be synthesized in a single set of linked reactions. Inorganic phosphate serves as both a general acid/base catalyst (HA/A- in the figure below) in these new reactions in the formation of an important intermediate, 2-amino-oxazole, and as a nucleophilic catalyst.
Figure: Abiotic Synthesis of 2-amino-oxazole
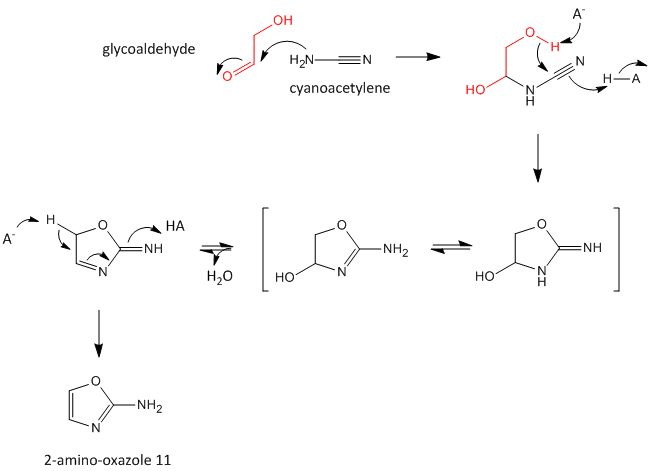
Glyceraldehyde, 2-amino-oxazole, and inorganic phosphate can react to form a ribocytidine phosphate. Possible reaction mechanisms are shown below.
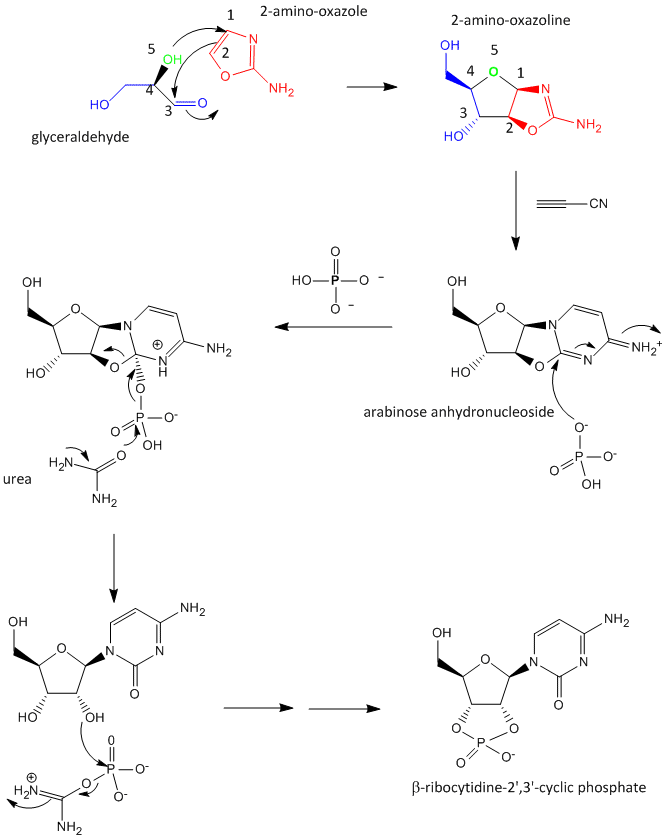
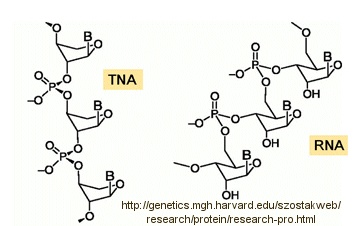
Abiological synthesis of polymer precursors is a long way from creating genetic polymers like RNA and DNA. These genetic polymers have one property that at first glance seems not conducive to a genetic molecule. Both are polyanions, which must be packed into a cell and folded onto itself to form the classic dsDNA helix and many different RNA structures. This problem is solved to some degree by the presence of counterions that help mask the charge on the negative backbone of the nucleic acids. The presence of phosphate in the phospodiester backbone linkage does confer an important advantage over other possible links (carboxylic acid esters, amides and anhydrides). The electrophilic phosphorous atom is hindered from nucleophilic attack by the negative O attached to the phosphorous. Also, the phosphorous is sp3 hybridized compared to the sp2 hybridization of the electrophilic carbon atom in anhydrides, esters, and amides, and hence is less accessible to nucleophilic attack. Most people now believe that RNA, which can act both as an enzyme and genetic template, preceded DNA as the genetic carrier. The evolution of DNA as the primary genetic carrier required an enzyme to convert ribose to deoxyribose. This would make the nucleic acid less likely to cleave at the phosphodiester bond with the replacement of a nucleophilic 2' OH with an H, and make the genetic molecule more stable. Other types of genetic carriers might have preceded the RNA world, especially if the monomer required could be more readily synthesized from abiological sources. One such alternative are threose nucleic acids (TNA). Synthetics ssTNA can base pair with either RNA, DNA, or itself to form duplexes.
![]() http://genetics.mgh.harvard.edu/szostakweb/research/protein/research-pro.html
http://genetics.mgh.harvard.edu/szostakweb/research/protein/research-pro.html
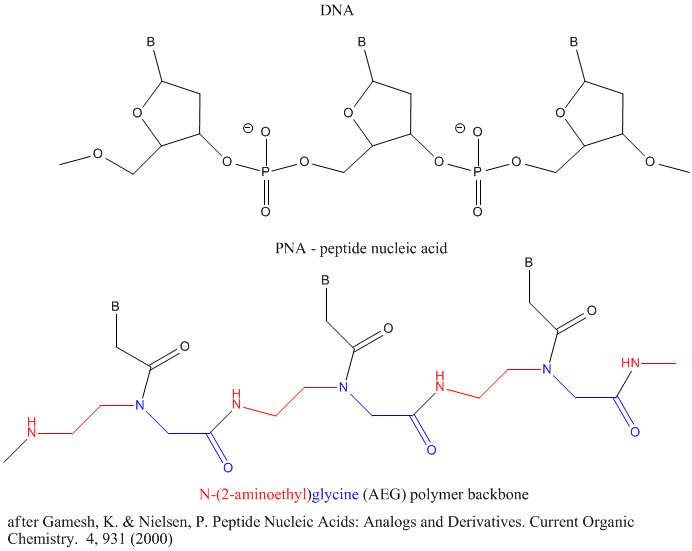
Other possible candidate include peptide nucleic acids (PNA). These can also form double stranded structures with DNA, RNA, or PNA single strands. They were initially designed to bind to dsDNA in the major grove forming a triple-stranded structure. Binding could alter DNA activity, possibly by inhibiting transcription, for example. The structure of a single-stranded PNA is shown. Note that the backbone, a polymer of N-(2-aminoethyl)glycine (AEG) which can be made in prebiotic soups, is not charged, making it easier to bind to dsDNA. AEG polymerizes at 100oC to form the backbone.
In addition to changing the backbone, additional bases other than A, C, T, G, and U can be accommodated into dsDNA and ssRNA molecules (Brenner, 2004)
In a recent extension, Pinheiro et al have shown that 6 different foreign backbone architectures can produce xeno-nucleic acids (XNAs) that can be replicated by engineered polymerases which make XNAs from a complementary DNA strand, and a polymerase that can make a complementary copy of DNA from an XNA. XNAs can also be evolved as aptamers to bind specific target molecules. The investigators replaced the deoxyribose and ribose backbone sugar with xenoanalogs (congeners) including 1,5-anhydrohexitol (HNAs), cyclohexene (CeNA), 2'-O,4'-C-methylene-b-D ribose (locked nucleic acids - LNA), L-arabinose (LNA), 2'-fluoro-L-arabinose (FANAs) and threose (TNAs) as shown in the figure below.
Figure: Xeno-nucleic acid sugar congeners
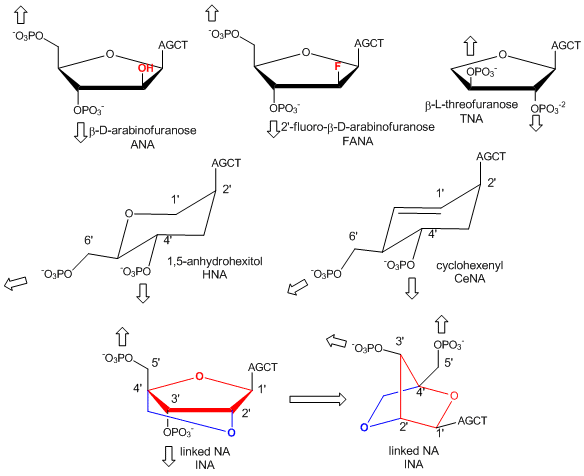
Polymers of these XNA can bind to complementary RNA and DNA and as such act as nuclease-resistant inhibitors of translation and transcription.
Von Kiedrowski, in an experiment similar to the self-replication of peptides described above, has shown that a single stranded 14 mer DNA strand, when immobilized on a surface, can serve as a template for the binding of complementary 7 mers and their conversion to 14 mers. When released by base, this process can occur with exponential growth of the complementary 14 mers. (von Kiedrowski Nature, 396, Nov 1998). Ferris has shown that if the clay montmorillonite is added to an aqueous solution of diadensosine pyrophosphate, polymerization occurs to produce 10 mers which are 85% linked in a 5' to 3' direction.
There are other reasons why polyanions are useful genetic molecules, other than their resistance to nucleophilic attack. The biological form of DNA is a large double stranded polyanionic polymer, in contrast to RNA which is a single-stranded polyanion polymer and protein which are polymers with varying combination of anionic, cationic, and hydrophobic properties. Even with counterions, it would be difficult to fold DNA into complicated and compact 3D structures as occurs for proteins, given the large electrostatic repulsions among the charged phosphates. Rather it forms a elongated double stranded rod, not unlike the rod-like structure of proteins denatured with sodium dodecyl sulfate (used in SDS PAGE gels). The elonged rod-shaped structure of ds-DNA is critical for the molecule which is the main carrier of our genetic information since mutations in the bases (leading to a switch in base pairs) causes no change in the overall structure of dsDNA. This enables evolutionary changes in the genetic material to produce new functionalities. A single change an amino acid of a protein, however, can cause a large change in the structure of a whole protein, a feature unacceptable for a carrier of genetic information. RNA structure effectively lies between that of DNA and proteins. Since it has less charge density than dsDNA, it can actually form dsRNA helices, so it can carry genetic information, as well as form complex 3D shapes necessary for its activity as an ribozyme. Perhaps more importantly, steric interference prevents ribose in RNA from adopting the 2'endo conformation, and allows only the 3'endo form, precluding the occurrences of extended ds-B-RNA helices.
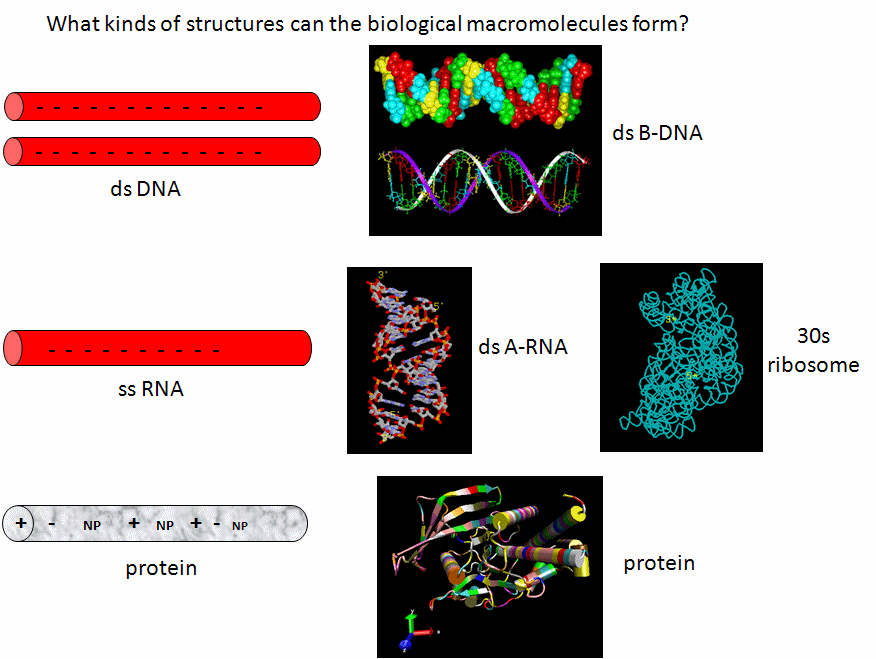
Let's assume that abiological precursors would react to form polymer-like molecules that might be complex enough to fold to structures that would allow binding, catalysis, and rudimentary replication. All this would be worthless unless they could be sequestered in a small volume which would limit diffusion and increase their local concentration. What is required is a membrane structure. Amphiphilic molecules, like lipids, with which we started this book, would be prime candidates since they spontaneously assembly to form micelles and bilayers, as shown in the review diagram below.
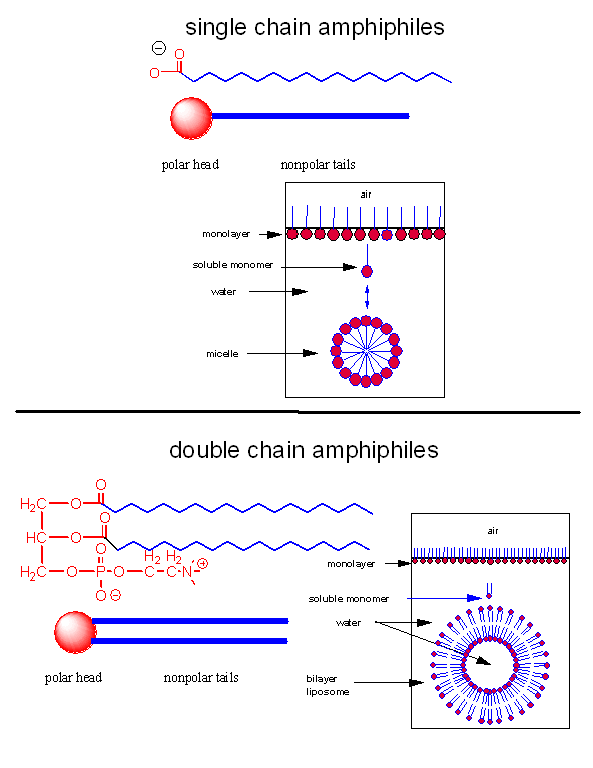
As mentioned in Chapter 1, alternative lipid phases are possible. Bilayers can also be formed from single chain amphiphiles, such as certain fatty acids, as illustrated in the equilibrium shown below. This occurs more readily at pH values close to the the pKa of the fatty acid, at which the fatty acids are not all deprotonated with full maximal negative charges. Single chain amphiphiles like fatty acids, which were more likely to formed in abiotic conditions, have been found in meteorites.
Clay surfaces, which have been shown to facilitate the formation of nucleic acids polymers, can also promote the conversion of fatty acid micelles to bilayers (Szostak). One such clay surface, montmorillonite, whose structure is shown below, promotes bilayer formation. It has an empirical formula of Na0.2Ca0.1Al2Si4O10(OH)2(H2O)10.
![]()
![]() Chime
Molecule Modeling: montmorillonite
Chime
Molecule Modeling: montmorillonite ![]()
![]() : montmorillonite
: montmorillonite
The effect of montmorillonite on vesicle formation can be shown by simple measurements of turbidity with time. Microscopy of fluorophore-encapsulated vesicles also shows encapsulated montmorillonite. The fatty acids presumably absorb to the cation layer of the clay particles. Time studies using light scattering also indicate that the vesicles grow in the presence of fatty acid micelles. To differentiate between the formation of new vesicles and the increase in size of pre-existing vesicles (which couldn't be done by simple light scattering without separation of the vesicles), investigators used two different fluorescent molecules to label fatty acid vesicles. The two probes were selected such that if the two probes came in close contact, energy transfer from the excited state of one fluorophore to the other fluorophore could occur, an example of fluorescence resonance energy transfer (FRET). FRET is observed when emission of the second dye occurs after excitation of the first dye, at a wavelength outside of the excitation wavelength of the second dye. If unlabeled vesicles were added to either labeled vesicles, no changes in FRET were observed, suggesting that the dyes did not move between vesicles. If fatty acid micelles were added, a decrease in FRET was observed, suggesting that new fatty acids were transferred to the doubly-labeled vesicles, effectively diluting the dye concentrations in the bilayer and their relative proximity, both which would decrease FRET. Most of the new fatty acid was incorporated into pre-existing vesicles which grew. The vesicles could also divide if extruded through a small pore. Later we will see that the energy to grow the vesicles can derive in part from a transmembrane proton concentration collapse. Division of vesicles might be promoted by bilayer assymetries associated with addition of substances to the outer leaflet, causing membrane distortion.
At some point, early genetic material must have been encapsulated in a membranous vesicles. Would new properties emerge from this mixture that might have a competitive (evolutionary) advantage over either component alone, and thus be a step on the way to the formation of a "living" cell? The answer appears to be yes. Chen et al. have incorporated RNA into fatty acid vesicles with interest effects. They asked the question as whether those vesicles could grow at the expense of vesicles without encapsulated RNA. RNA, with a high charge density and its associated counter ions would create osmotic stress on the vesicles membranes. To relieve that stress they could acquire fatty acids from other fatty acid vesicles (or fatty acid micelles), increasing their surface area, and concomitantly reducing tension in the membrane.
Oleic acids vesicles were first placed under stress by encapsulating 1 M sucrose in the vesicle and then diluting it in hypotonic media. Water would enter and swell the vesicle (but without bursting and resealing, as evident from control experiments). Then they prepared stressed and unstressed oleic acid vesicles in the presence of two nonpolar flurophores, NBD-PE (excitation at 430 nm, emission at 530 nm) and Rh-DHPE (emission at 586 nm). These fluorophores were chosen for fluorescence resonance energy transfer measurements. If the membrane vesicles changed size, the FRET signal would change, based on the relative concentration and proximity of the dual fluorophores. If the separation between probe molecules increased, the FRET signal would decrease. Conversely, if the vesicle shrunk, the FRET signal would increase.
The results showing the effect of adding unlabeled swollen vesicles to labeled normal vesicles, and labeled swollen vesicles to unlabeled normal are shown below. The surface area of normal labeled vesicles decreased by about 25% when unlabeled swollen vesicles were added, but not when unlabeled normal vesicles were added. Labeled swollen vesicles increased 25% in size only if mixed with unlabeled normal vesicles, not with unlabeled swollen vesicles. Hence swollen vesicles win the competition and "steal" lipid from normal vesicles.
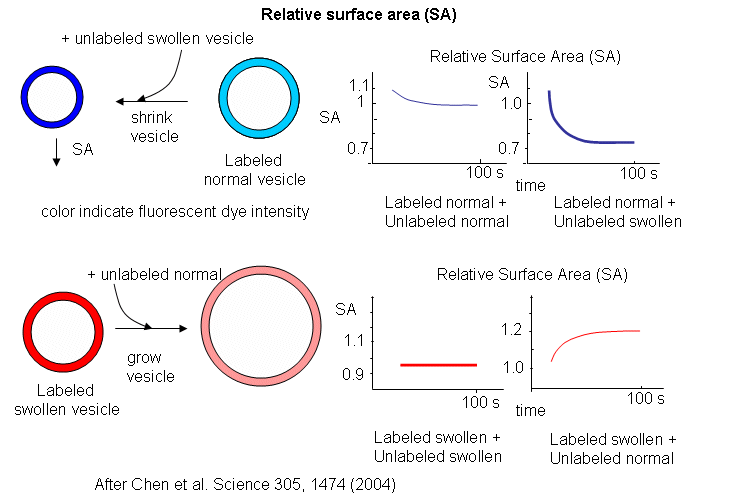
Now what about vesicles swollen with encapsulated RNA? RNA, with its associated charge and charged counter ions also placed an osmotic stress on the vesicles. FRET labels (the two fluorophores) were place in vesicles without RNA. Fatty acids were removed from isotonic labeled vesicles in the presence of unlabeled tRNA swollen vesicles (left panel below). Labeled vesicles swollen with glycerol took fatty acids from unswollen vesicles (without tRNA), but not from tRNA swollen vesicles, as both were swollen so no net drive to reduce swelling by lipid exchange was present.
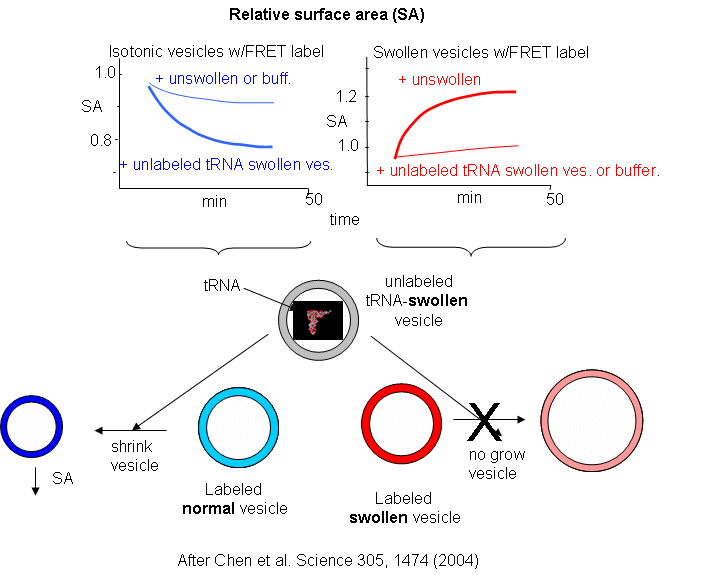
These results show the vesicles with encapsulated RNA have a competitive (evolutionary) advantage over normal vesicles. This data suggests that having a polyanion as the source of genetic material is actually advantageous to the protocell. In addition the move in modern membranes to phospholipids with esterified fatty acids (instead of free ones) may actually have stabilized membranes, given the movement of free fatty acids to different membranes.
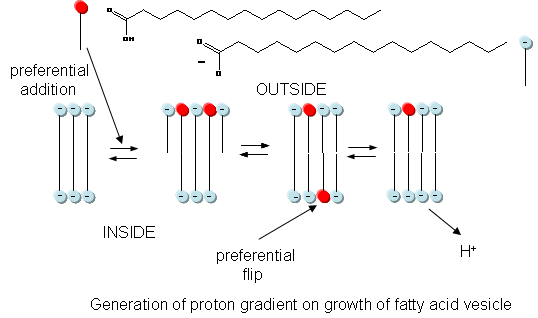
In addition to a genetic macromolecule and a semipermeable membrane, a source of energy to drive intracellular processes must be present. A common source of free energy used in many cells to drive unfavorable processes is a proton gradient, whose formation in modern cells can be coupled to energy input from oxidation, ATP cleavage, light, or the collapse of another gradient. Could a proton gradient be formed in protocells? It can, quite easily, when coupled to the growth of fatty acid vesicles. If a fatty acid vesicle is to grow, more fatty acid must be added to the outer leaflet. The protonated, uncharged form of the fatty acid would preferentially be added, since it would lead to less electrostatic repulsion between adjacent head groups. The protonated, uncharged form of the fatty acid would also be most likely to flip to the inner leaflet to minimize stress asymmetries in the leaflets. Once in the inner leaflet, it could deprotonate to form H+(aq) in the inside of the membrane, creating a transmembrane proton gradient and transmembrane potential. The energy released on growth of the membrane is partly captured in the formation of a proton gradient, as shown in the figure below.
The proton gradient would soon inhibit its own formation since further movement of protons into the cell would be attenuated by the positive transmembrane potential unless metal ions inside moved outside. In addition, the gradient would collapse after growth stopped. The investigators made fatty acids vesicles in the presence of pH 8.5 buffers whose pH was adjusted with an alkali metal hydroxide. The external pH was reduced to 8.0, resulting in a 0.5 pH unit proton concentration gradient. (Changes in intravesicular pH were measured with pH-sensitive fluorophore, HPTS.) Inward movement of protons down a concentration gradient, as shown in the figure below, would occur with time, collapsing the imposed concentration gradient.
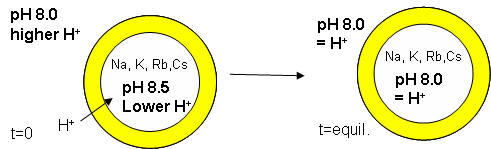
With fatty acid vesicles, this artificial pH gradient collapsed quickly, suggesting the vesicle permeability to protons was high. The rate was too high for simple flip-flop diffusion. Inward movement of protons appeared to be facilitated by outward movement of the M+ ions. The rate of decay of the proton gradient was exponential, and the resulting first order rate constant was easily determined. A graph of the rate constant for pH gradient collapse vs unsolvated ionic radius of M+ decreased with increasing radius (i.e. kNa > kK > KRb > KCs, suggesting that the pH gradient would be more stable if large, impermeable or otherwise trapped cations were encapsulated. When vesicles were made with encapsulated Arg+, the imposed pH gradient did not collapse for hours. If oleic acid micelles were added to oleic acid vesicles with encapsulated Arg+, with no artificial pH gradient induced across the membrane, the vesicle grew with concomitant movement of protons into the vesicle, producing a pH gradient of 0.3 within seconds.
These experiments show that membrane growth and energy storage could be coupled, and the right composition of encapsulated material could lead to a stable transmembrane pH gradient, a source of energy to drive biological processes. It even suggests that a charge polyanion would be beneficial as a genetic carrier.
The case for the origin of life in deep sea hydrothermal vents and not in a primordial "Campbell's" soup has been argued convincingly by Lane et al (2010). What's needed for life are reasonably complex molecules and an energy source to drive unfavorable reactions. It's the latter on which that Lane et al focus. In an early primordial world that was low in oxygen, exergonic oxidation reaction of organic molecules would provide little energy. This can be surmised from the low energy yield (compared to aerobic respiration) achieved in present day glycolytic (fermentative) pathways from all major domains of life, archaea, bacteria, and eukaryotes.
|
Background: Based on rRNA sequences, a primordial cell evolved into two different types of cells, one that became bacteria, and another that split further into archaea (single cells, similar to bacteria) and eukaryotic cells (complex cells with internal organelles that eventually formed multicellular organisms). Bacterial and archaea are collected called prokaroytic cells. |
In addition, these pathways required the evolution of up to dozen different enzymes to produce their relatively meager energy yield which ultimately depends on the oxidation of an organic molecule by another organic molecule instead of by a powerful oxidant like dioxygen. An anisotropic arrangement of molecules in a concentrated soup could lead to transient chemical potential fluctuations but these would be inefficient and impermanent sources of energy. Effectively the primordial soup would be at equilibrium and hardly expected to provide the energy for synthesis of RNA enzymes and replicators. UV light leads to photo-damage and photolysis not replication of complex molecules. What is needed is a way to drive the synthesis of molecule with high chemical potential energy (like sulfur esters and phosphoanhydrides) compared to their lytic products. These could then provide an energy sources to drive ATP synthesis, for example.
A detailed look a the bioworld shows that the earliest organisms used energy from the collapse of the proton gradient (chemisomotic principle elucidated by Peter Mitchell). All present autotrophs (organisms that can fix CO2 and form complex organic molecules) and many heterotrophs (use complex organic molecules of other organisms for fuel) use redox complexes in membranes coupled to membrane gradients. These complexes would take reduced molecules and pass electrons from them to oxidizing agents (electron acceptors), including O2, CO2, and Fe3+ to form H2O, CH4, and Fe2+. Fermentors also use ATPase membrane enzymes to transport nutrients. Yet genomic analysis of bacteria and archaea show that enzymes involved in fermentation differ significantly, suggesting that they evolved separately towards a convergent function. Structure in common include DNA, RNA, ribosomes and membrane ATPases, which Lane et al suggest were in a the Last Universal Common Cell (LUCA).
All autotrophs produce their energy source by fixing CO2 using either H2 directly or indirectly using H2O and H2S. All of the are available in nonhydrothermal deep sea vents. Volcanic vents, however, are extremely hot (not optimal for organic molecule synthesis), very acidic, and lack hydrogen gas. A different type of nonvolcanic vent, an alkaline hydrothermal one, might produce more conducive as a site of the origin of life. In these vents, water chemically reacts with minerals in the crust (such as olivine) leading to their hydroxylation and subsequent fracture, with promotes more water entry into the crust. It has been reported that there is more water found as hydroxylated minerals in the crust, that there is liquid water in the oceans. These processes result in temperatures up to 200 degrees Celsius and release of hydrogen gas into a moderately alkaline vents into the sea water at temperatures more conducive (70 degrees C) to the origin of life.
Figure: Alkaline Vent

Of the five different pathways known to fix CO2, all require ATP except one. That one is present in both methanogen, which produce methane from CO2 and H2 and in the acetogens, which produce acetate (CH3CO2-) in the form of acetylCoA. The simpler reactions of forming acetic acid and methane are shown below:
2CO2 + 4H2 --> CH3CO2H + 2H2O.
CO2 + 4H2 --> CH4 + 2H2O.
The DG0 values for these reaction (calculated using DG0f for gas phase H2, CO2 and CH4 and liquid acetic acid and water are -75 and -131 kJ, respectively, at 250C, showing that they are thermodynamically favored. Making AcetylCoA, a "high" energy molecule compared to its hydrolysis products (as is ATP) from acetic acid and CoASH, a would require energy input. A proton gradient is the likely source.
Some bacteria and Achaea cells (primordial or present) use the reductive acetylCoA pathway, also known as the Wood-Ljungdahl pathway, to form, in a noncyclic process, acetyl CoA from CO2 and at the same time makes ATP. This process is paid for by a proton gradient. This has been described by Shock as "a free lunch you get paid to eat". The energetics of the present acetyl CoA pathway based on the overall reaction below show an approximate DG0 value of -59 kJ/mol which can drive ATP synthesis.
2CO2 + 4H2 + CoASH --> CH3COSCoA + 3H2O.
The concentration of carbon dioxide in the primordial ocean was 1000 times higher than now. Vents produced large amounts of methane and hydrogen gas. There was little oxygen and hence lots of Fe2+. The enzymes involved in this acetyl-CoA pathway of carbon fixation have FeS clusters. It has also been shown that bubbles (which are really membrane bound spaces) of FeS and NiS can be made in deep sea vents. These could not only encapsulate precursor molecules but also serve as catalysts. Vents also can catalyze the fixation of nitrogen (to ammonia) and laboratory studied show that FeS can catalyze the conversion of formate (found in vents) into pyrimidines and purines. The studies of present methanogens and vent chemistry suggest that the critical ingredients and conditions for development of the first biological cells probably occurred in the vents.
To produce polymers, an energy source and monomers must exists. Concentration gradients found in simulations of vents produce million fold concentrated molecules. The transient heating and cooling of any double-stranded nucleic acids could lead to concentration amplification by a PCR like strand separation followed by reannealing. In addition, these vent regions possess a powerful, reoccurring energy source, a pH gradient, as the alkaline vented material entered the acidic oceans that exists with high CO2 concentrations, creating a gradient across an inorganic membrane. This is startlingly analogous to the pH gradient across membranes (acidic outside, alkaline inside) driven by the membrane complexes in the mitochondria and bacteria. Lane et al argue that the existence of membrane proton gradients as an energy source in all cells (eukaryotes, bacteria, and archaea) and in chloroplasts, mitochondria, corroborate their hypothesis. Bacteria and archaea share homologous ATPases and electron carriers (ferredoxins, quinones, and cytochromes). These similarities contrast to the differences in enzyme structures in fermentative pathways. Arguments that proton pumps evolved to pump proteins (and reduce pH gradients) can't explain their ubiquitous presence even in organisms not subjected to low pH. Hence the ubiquity of proton pumps supports the conjecture that they arose from the first protocells, possible comprised of inorganic walls and ultimately with amphiphilic molecules synthesized from precursors.
Creationists would argue that it would be impossible to evolve a structure with the complexity of membrane ATPase (which serve to collapse a pH gradient as the power the synthesis of molecules with large negative DG0 of hydrolysis). Lane et al propose that the earliest cells evolved ATPase like molecules in alkaline vents where pH gradients analogous to those in cells today arose. They envision cell-like columns lined by FeS membrane like structure with alkaline conditions inside and acid conditions outside. Nonpolar or amphiphilic molecule would line the inside of the cells/columns. A ATPase-like system could then take advantage of the pH gradient which constantly replenishes itself. If structures as complicated as ribosomes evolved from a subsequent RNA world, surely ATPase-like molecules could also. Other chemistry might have evolved earlier to utilize the energy source provided by the pH gradient.
If life originated in the vents, it would need an energy source to leave the vents. Presumably it would have evolved one to utilized pH gradient to replace the one it left in the alkaline vents. The substrate level phosphorylation of glycolysis that requires ATP input to make ATP would not provide the energy source needed. Cells that left would have had to produce their own proton gradient. Perhaps all the was needed initially was concerted conformational changes in proteins that upon exposure of a different pH changed their shape inducing pKa shifts in adjacent proton donors/acceptors leading to vectorial discharge of protons across a membrane. Perhaps the method described above in protocells was sufficient.
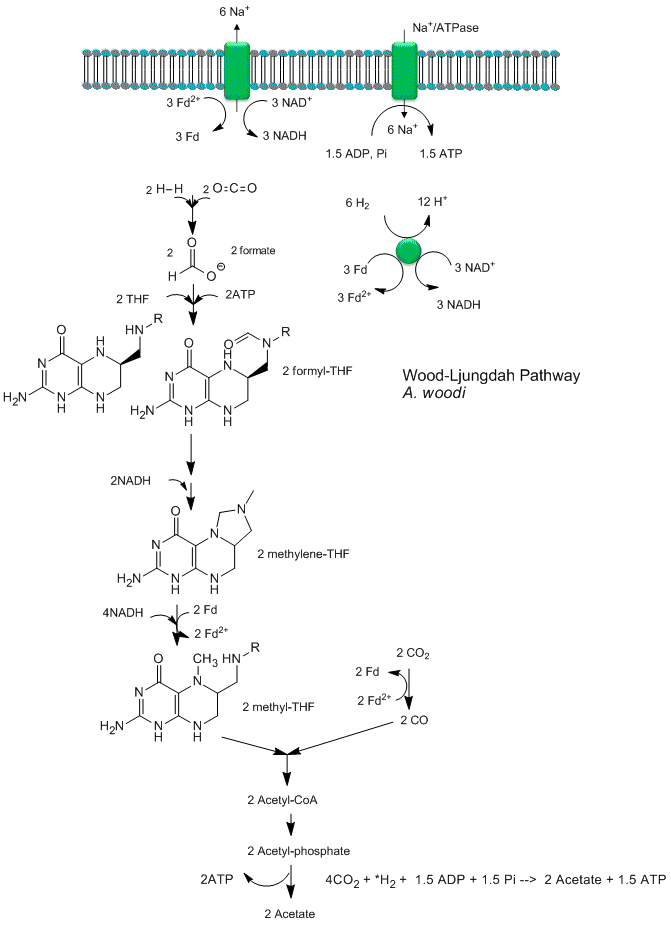
Recent analyses by Poehlein et a show that CO2 reduction (fixation) can be coupled to the production of a sodium ion gradient, which could collapse to drive ATP synthesis. Analysis of the genome of a gram positive bacteria, Acetobacterium woodii, an acetogen, shows the it has an ancient pathway for production of acetyl-CoA that can, in an anabolic fashion form biomass or in a catabolic fashion be cleaved to acetate with the production of ATP. It does not require classic electron carriers like ubiquinone or cytochrome C linked to protein gradient formation to drive ATP synthesis. Rather it has only a ferredoxin:NAD+ oxioreductase which couples oxidation to the formation of a sodium ion gradient, which collapses through an sodium ion transporter/ATP synthase to drive ATP synthase. A plausible reaction scheme based on genomic analysis is shown below:
Figure: Acetyl-CoA Synthase and Acetogenesis
Let's return to the chicken and egg dilemma one more time. What is needed for biological polymer formation are monomeric precursors, an energy source, and a way to compartmentalized them all. We discuss how monomeric precursors could form, but wouldn't it be far better if even the synthesis of precursors could be catalyzed? One source of catalysis mostly absent from the "bioorganic" abiotic chemistry in the above discussion is the transition metals. Transition metals can form complexes. Ligands containing lone pairs on O, N, and S atoms can donate them to transition metals ions, which can hold up to 18 electrons in s, p, and d orbitals. Hence as many as 9 lone pairs on ligand molecules (which are often multidentate) could be accommodated around the transition metal ion. Many present small molecule metabolites and their abiotic precursors (H2O, CO, CO2, NH3 and thiols) bind cations as mono- or polydentate donors of electrons. Hence transition metal ions would have a thermodynamic tendencies to be bound in complexes.
Bound ligands that contain potentially ionizable hydrogens could become deprotonated and made better nucleophiles for reactions. Hence the transition state metal ion, acting with the complex, becomes a catalyst as it decreases the pKa of a bound ligand (such as water). In addition, since transition metals ions can have multiple charge and oxidation states, they can easily act as redox centers in the oxidation/reduction of bound ligands that were redox active. Given the relative anoxic conditions of the early oceans, Fe2+ would predominant. It could easily be oxidized to Fe3+ as it reduced a bound ligand. Highly charged transition states would withdraw electron density from bound ligands leading to their possible oxidation.
Metals obviously still play a strong role in catalysis, both indirectly in promoting correct protein folding and directly in stabilizing charge in both the transition state and intermediates in chemical reaction pathways. FeS clusters are of significant importance. Their biosynthesis involves removal by an active site Cys in a desulfurase enzyme of a sulfur from a free amino acid Cys followed by its transfer to an Fe in a growing FeS cluster in a FeS scaffold protein, which then transfers the cluster to an acceptor protein where it acts as a cofactor. FeS clusters can adopt a variety of stoichiometries and shapes, as well as redox states for the participating Fe ions. The continuing importance of FeS clusters in all cells, their involvement in not only redox enzymes in which electron transfer is facilitated by delocalization of electrons over both Fe and S centers, but also in coupled electron/proton transport in mitochondrial electron transport, Fe storage (ferrodoxins), and in regulation of enzyme activity and gene expression, suggests that they were of primordial importance in the evolution of life. T
hey are often found at substrate binding sites of FeS enzymes involved in both redox and nonredox catalysis. A ligand can bind to a particular Fe in the cluster, activating it for hydration or dehydrogenation reactions. Fe 4 of the FeS cluster in the TCA enzyme aconitase can have a coordination numbers of 4, 5, or 6 as it binds water, hydroxide or substrate. It acts to both decrease electron density in the transition state and to change the pKa of bound water as the enzyme catalyzes an isomerization of tricarboxylic acids (citric and isocitric acid) through an elimination/addition reaction with water. In another example it can bind S-adenosylmethionine through its amine and carboxylate groups, which activates the molecule for cleavage and radical formation. In some cases metals other than Fe (Ni for example) are incorporated into the cluster. FeS effects on transcription factors involves facilitation of optimal structure for DNA binding. FeS and FeNi centers in proteins are similar in structure tp FeS units in minerals like greigite and presumably to FeS structure formed when H2S and S2- react with Fe2+ (present in abundance in the early ocean) and other metals in vents Metal sulfides participate in reduction of both CO and CO2. For example the synthesis of CH3SH from CO2 and H2S is catalyzed by "inorganic" FeS.
This question is being addressed by eliminating "unnecessary" gene from simple bacteria. Cells placed in a rich nutrient broth with essential lipids, vitamins, and amino acids would need fewer genes than those placed in a more nutrient-poor medium. Bacteria cells like Mycoplasma genetalium, that live within "nutrient rich" eukaryotic cell, have been genetically manipulated to delete unnecessary genes. Based on knockout studies, it may be possible for the cell to survive with only 300-350 genes. Bacillus subtilis has approximately 4100 genes. Estimates have been made that it could survive with as few as 271 genes.
more to be added
![]() The
Origins of Life on Earth, Leslie Orgel
The
Origins of Life on Earth, Leslie Orgel
![]() Venter
synthesizes whole bacterial genome |
Venter
synthesizes whole bacterial genome |
![]() Ted
Talk:
Protocells
Ted
Talk:
Protocells
References
Longo, L, Lee, Jihun and Blaber, M. Simplified protein design biased for prebiotic amino acids yield a foldable, halophilic protein. PNAS. 110, 2135 (2013)
Morowitz, H. et al. Ligand Field Theory and the Origin of Life as an Emergent Feature of the Periodic Table of Elements. Biol Bull. 219, 1-6 (2010)
Pinheiro, V. et al. Synthetic genetic polymers capable of heredity and evolution. Science 336, 341 (2012)
Lane, N et al. How did LUCA make a living? Chemisomosis in the origin of life. BioEssays 9999, 1-10 (2010) DOI 10.1002/bies.200900131
Russell, M, and Martin, W. The rocky roots of the AcetylCoA pathway. TIBS, 29, pg 358 (2004) doi:10.1016/j.tibs.2004.05.007
Shock EL, McCollom TM, Schulte MD. The emergence of metabolism from within hydrothermal systems. In Wiegel J, Adams MWW. ed; Thermophiles: The Keys to Molecular Evolution and the Origin of Life. London: Taylor & Francis. p 59�76.a998)
Powner, M et al. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459, 239-242 (14 May 2009) | doi:10.1038/nature08013;
Chen, I. The Emergence of Cells During the Origin of Life. Science 314, 1558 (2006)
Chen, I., Roberts, R. & Szostak, J. The emergence of competition between model protocells. Science 305, 1474 (2004)
Chen, I and Szostak, J. Membrane growth can generate a transmembrane pH gradient in fatty acid vesicles. PNAS, 101, 7965 (2004)
Leman, L, Orgel, L, and Ghadiri, M. Carbonyl Sulfide-Mediated Prebiotic Formation of Peptides. Science 306, 283 (2004)
Brenner, S. Understanding Nucleic Acids Using Synthetic Chemistry. Acct. Chem. Research 37, 784 (2004)
Hanczyc, J., Fukikawa, S. and Szostak, J. Experimental Models of Primitive Cellular Compartments: Encapsulation, Growth, and Division. Science 302, 618 (2003)
Kobayashi, K. et al. Essential Bacillus subtilis genes. PNAS 100, 4678 (2003)
Rasmussen, S. et al. Bridging Nonliving and Living Matter. Artificial Life 9: 269 (2003)
Krishna N. Ganesh and Peter E. Nielsen. Peptide Nucleic Acids: Analogs and Derivatives. Current Organic Chemistry. 4, 931 (2000)
Saghatelian, A. , Yokobayashi Y, Soltani K, Ghadiri MR. A chiroselective peptide replicator. Nature 409, 797 (2001). Nelson, K., Levy, M. & Miller, S. Peptide nucleic acids rather than RNA may have been the first genetic molecule. PNAS 97, 3868 (2000)
Luther, A., Brandsch, R. & von Kiedrowski, G. Surface-promoted replication and exponential amplification of DNA analogues. Nature, 396, 245 (1998)
Ferris, J, & Ertem, G et al. Oligmerization of ribonucleotides on montmorillonite: reaction of the 5'-phosphorimidazolide of adenosine. Science. 257, 1387 (1992)
Navigation
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 10: The Origin of Life
