Biochemistry Online: An Approach Based on Chemical Logic

Structure & Reactivity in Chemistry
Metabolic Pathways
MP4. Regulation of Metabolic Pathways A: How is enzyme activity regulated?
Henry Jakubowski
Exquisite mechanisms have evolved that control the flux of metabolites through metabolic pathways to insure that the output of the pathways meets biological demand and that energy in the form of ATP is not wasted by having opposing pathways run concomitantly in the same cell.
Enzymes can be regulated by changing the activity of a preexisting enzyme or changing the amount of an enzyme.
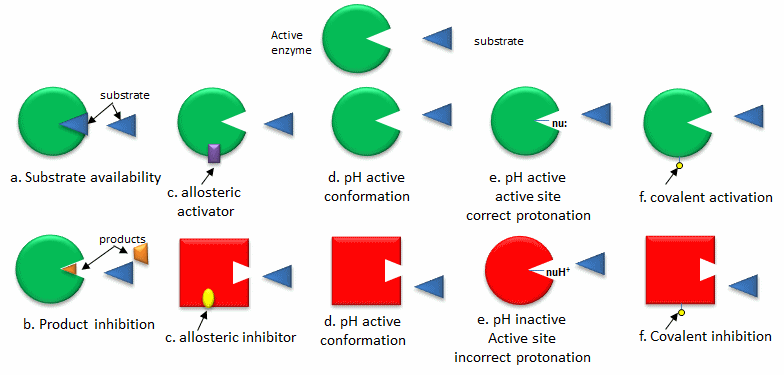
A. Changing the activity of a pre-existing enzyme: The quickest way to modulate the activity of an enzyme is to alter the activity of an enzyme that already exists in the cell. The list below, illustrated in the following figure, gives common ways to regulate enzyme activity
- Substrate availability: Substrates (reactants) bind to enzymes with a characteristic affinity (characterized by a dissociation constant) and a kinetic parameter called Km (units of molarity). If the actual concentration of a substrate in a cell is much less than the Km, the activity of the enzyme is very low. If the substrate concentration is much greater than Km, the enzyme active site is saturated with substrate and the enzyme is maximally active.
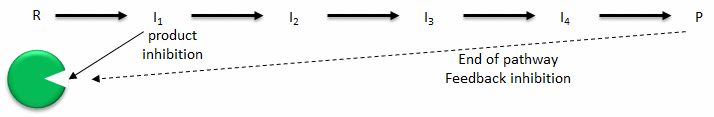
- Product inhibition: A product of an enzyme-catalyzed reaction often resembles a starting reactant, so it should be clear that the product should also bind to the activity site, albeit probably with lower affinity. Under conditions in which the product of a reaction is present in high concentration, it would be energetically advantageous to the cell if no more product was synthesized. Product inhibition is hence commonly observed. Likewise it be energetically advantageous to a cell if the end product of an entire pathway could likewise bind to the initial enzyme in the pathways and inhibit it, allowing the whole pathway to be inhibited. This type of feedback inhibition is commonly oberved
- Allosteric regulation: As many pathways are interconnected, it would be optimal if the molecules of one pathway affected the activity of enzymes in another interconnected pathway, even if the molecules in the first pathway are structurally dissimilar to reactants or products in a second pathway. Molecules that bind to sites on target enzymes other than the active site (allosteric sites) can regulate the activity of the target enzyme. These molecules can be structurally dissimilar to those that bind at the active site. They do so my conformational changes which can either activate or inhibit the target enzyme's activity.
- pH and enzyme conformation: Changes in pH which can accompany metabolic process such as respiration (aerobic glycolysis for example) can alter the conformation of an enzyme and hence enzyme activity. The initial changes are covalent (change in protonation state of the protein) which can lead to an alteration in the delicate balance of forces that affect protein structure.
- pH and active site protonation state: Changes in pH can affect the protonation state of key amino acid side chains in the active site of proteins without affecting the local or global conformation of the protein. Catalysis may be affected if the mechanism of catalysis involves an active site nucleophile (for example), that must be deprotonated for activity.
- Covalent modification: Many if not most proteins are subjected to post-translational modifications which can affect enzyme activity through local or global shape changes, by promoting or inhibiting binding interaction of substrates and allosteric regulators, and even by changing the location of the protein within the cell. Proteins may be phosphorylated, acetylated, methylated, sulfated, glycosylated, amidated, hydroxylated, prenylated, myristolated, often in a reversible fashion. Some of these modifications are reversible. Regulation by phosphorylation through the action of kinases, and dephosphorylation by phosphates is extremely common. Control of phosphorylation state is mediated through signal transduction process starting at the cell membrane, leading to the activation or inhibition of protein kinases and phosphatases within the cell.
Figure: Regulation of the Activity of Pre-existing Enzymes
Extracellular regulated kinase 2 (ERK2), also known as mitogen activate protein kinase 2 (MAPK2) is a protein the plays a vital role in cell signaling across the cell membrane. Phosphoryation of ERK2 on Threonine 183 (Thr153) and Tyrosine 185 (Tyr185) leads to a structural change in the protein and the regulation of its activity.
Jmol: Erk2 -Structural Comparison of phosphorylated and dephosphorylated enzyme
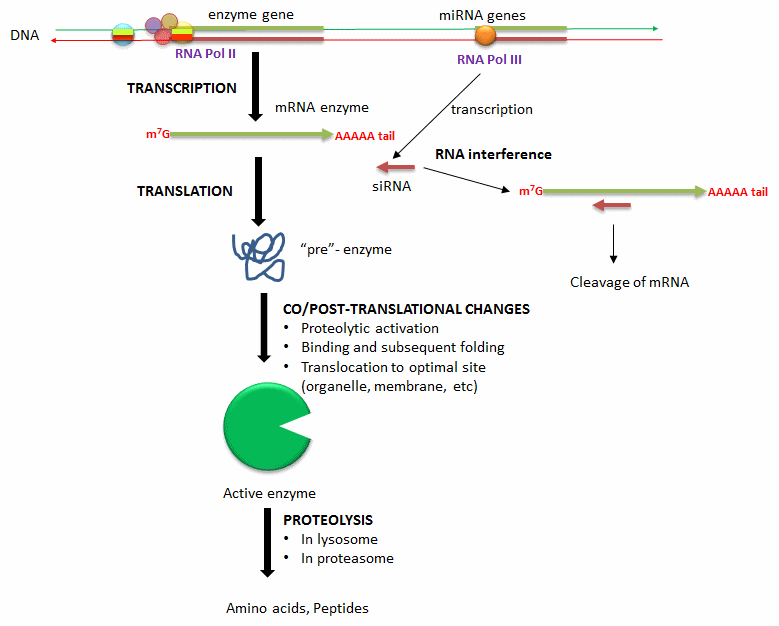
B. Changing the amount of an enzyme: Another and less immediate but longer duration method to modulate the activity of an enzyme is to alter the activity of an enzyme that already exists in the cell. The list below, illustrated in the following figure, shows way in which enzyme concentration is regulated.
- Alternation in transcription of enzyme's gene: Extracellular signal (hormones, neurotransmitters, etc) can lead to signal transductions responses and ultimate activation or inhibition of the transcription of the gene for a protein enzyme. These changes result from recruitment of transcription factors (proteins) to DNA sequences that regulate transcription of the enzyme gene.
- Degradation of messenger RNA for the enzyme: The levels of messenger RNA for a protein will directly determin the amount of that protein synthesized. Small inhibitor RNAs, derived from microRNA molecules transcribed from cellular DNA, can bind to specific sequences in the mRNA of a target enzyme. The resulting double-stranded RNA complex recruits an enzyme (Dicer) that cleaves the complex with the effect of decreasing translation of the protein enzyme from its mRNA.
- Co/Post translational changes: Once a protein enzymes is translated from its mRNA, it can undergo a changes to affect enzyme levels. Some proteins are synthesized in a "pre"form which must be cleaved in a targeted and limited fashion by proteases to active the protein enzyme. Some proteins are not fully folded and must bind to other factors in the cell to adopted a catalytically active form. Finally, fully active protein can be fully proteolyzed by the proteasome, a complex within cells, or in lysosomes, which are organelles within cells containing proteolytic enzymes.
Next we will consider which enzymes in pathways make the best target for regulation.
Navigation
| Reactivity Index | MP0. Metabolic Pathways - Main Menu |
| Structure & Reactivity Web Materials | Biochemistry Online: Table of Contents |

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.