Determination of Mechanism in Chemistry
Mathematical Tools in Kinetics
MK12. Determination of Bond Strengths
Bond strengths are fundamental parameters in chemistry. They influence the thermodynamics of reactions through their contributions to the energies of reactants and products. They influence the kinetics of reactions by contributing to activation barriers. These quantities are important to know.
How do we determine a bond strength? Furthermore, what do we mean by bond strength? There are different things that might come to mind when we think about breaking a bond. We might think of ionizing a bond, for instance, so that one atom gets the electrons in the bond and the other atom does not. That's called heterolytic cleavage.
A-B → A: - + B+
Alternatively, maybe we are thinking about homolytic cleavage. Rather than forming two ions, we break the bond in half to get two neutral species.
A-B → A. + B.
Those situations are very different. They would cost very different amounts of energy, partly because stabilization of an anion and cation wouldn't be the same as stabilization of a pair of radicals. So, the first thing we have to do is all get on the same page about what we mean when we talk about a bond strength. If you look up a table of bond strengths, what you are looking at is a measurement of the latter case, a homolytic bond cleavage to produce two radicals. The heading on the table frequently says "BDE" for bond dissociation enthalpy, which was the thermodynamic quantity that was measured in the majority of cases.
How do they measure these things? There are lots of ways. Conceptually, the simplest thing to do would be to measure an equilibrium constant between the covalent compound and its dissociated radicals. In some cases, people have essentially done that using mass spectrometry, although in that method they are actually observing cations formed from all three species. Remember, mass spectrometry involves the generation of cationic species that can be deflected by a magnetic field.
A-B+ → A.+ + B. (or A. + B.+)
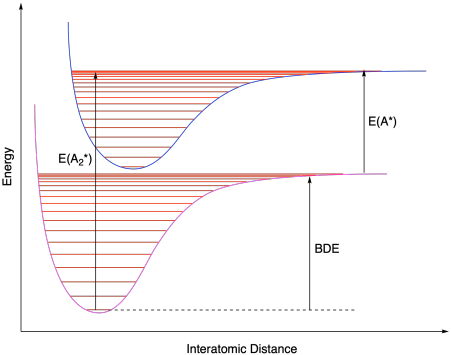
Bond strengths can also be measured in relatively simple diatomic molecules using spectroscopic methods. UV spectra of gas-phase halogens, for example, show fine structure that can be used to calculate the energy difference between the first vibrational level in the ground state and the highest vibrational level in the excited state. Coupled with spectroscopic evidence of the energy difference between the lowest and highest vibrational level in the excited state of a halogen atom, the bond dissocation energy can be deduced. There's a little more to it than that, but the bond dissociation enthalpy is essentially the difference, E(A2*) - E(A*) on the diagram below.

Figure MK12.1. Electronic ground state and excited states with corresponding vibrational levels.
Problem MK12.1.
Given the following experimentally-derived measurements for bromine, determine the strength of the halogen-halogen bond: E(A2*) = 19,898 cm-1; E(A*) = 7598 cm-1. Note that 1 cm-1 translates into 2.86 cal/mol.
(F.E. Stafford, Band spectra and dissociation energies. J. Chem. Educ. 1962, 39, 626-629.)
It might seem like we should have bond strengths all sorted out by now. Take a look at a water molecule using one of these methods and we've got the strength of an O-H bond. Take a look at a methane molecule and we have the strength of a C-H bond. It shouldn't take long to build up a library of bond strengths that would cover most molecules we might encounter.
Things are a little more complicated than that because a known bond strength in one molecule doesn't translate into the same bond strength in another molecule. Breaking an O-H bond in water, for example, requires about 110 kcal/mol, whereas an O-H bond in phenol only requires around 80 kcal/mol; that's an enormous percentage difference. The H3C-H bond in methane costs about 105 kcal/mol to break, but it only takes about 88 kcal/mol to break the CH2CHCH2-H bond in propene. There's a definite need to know more details about bond strengths in specific molecules, but larger molecules make the determination of an individual bond strength a little trickier.
Problem MK12.2.
Provide reasons for the very different bond strengths in these cases.
a) C6H5O-H vs. H2O (80 vs. 110 kcal/mol)
b) CH2CHCH2-H vs. CH4 (88 vs. 105 kcal/mol)
Bond strength determinations in slightly larger molecules usually invoke Hess' Law, deriving a specific bond strength by constructing an imaginary thermodynamic cycle to get from the species of interest to its two dissociated radicals. The simplest way to do that is through heats of formation. For example, we could compare the heats of formation of propene to the heats of formation of the two radicals that result when the primary C-H bond breaks.
Those heats of formation are associated with these reactions:
3 C(s) + 3 H2(g) → CH2=CH-CH3(g) ΔHf = 4.9 kcal/mol
3 C(s) + 2.5 H2(g) → CH2=CH-CH2(g) ΔHf = 40.9 kcal/mol
0.5 H2(g) → H(g) ΔHf = 52.1 kcal/mol
Which we can rearrange to get:
CH2=CH-CH3(g) → 3 C(s) + 3 H2(g) ΔH = -4.9 kcal/mol
3 C(s) + 2.5 H2(g) → CH2=CH-CH2(g) ΔHf = 40.9 kcal/mol
0.5 H2(g) → H(g) ΔHf = 52.1 kcal/mol
And summing up gives:
CH2=CH-CH3(g) + 3 C(s) + 2.5 H2(g) + 0.5 H2(g) → CH2=CH-CH2(g) + 3 C(s) + 3 H2(g) + H(g) ΔH = 93.0 -4.9 kcal/mol
or
CH2=CH-CH3(g) → CH2=CH-CH2(g) + H(g) ΔH = 88.1 kcal/mol
Heats of formation like these can often be found in thermochemistry data tables like those compiled by the National Institute of Standards and Technology (NIST). Alternatively, they can often be generated pretty easily using a reliable computational chemistry approach.
There are other ways of using thermodynamic cycles to get bond strength data. If the bond is also known to be acidic, we might construct a cycle more like this:
H-A → H+ + A-
H+ + e- → H.
A- → A. + e-
If we add those equations together, we get:
H-A + H+ + e- + A- → H+ + A- + H. + A. + e-
or
H-A → H. + A.
We just need appropriate energies that correspond to each of those reactions. The first one (H-A → H+ + A-)could be determined by measuring pKa, because of the relationship between free energy and the equilibrium constant (ΔG = -RTlnK). The second one (H+ + e- → H.) could be obtained from the ionization energy of a hydrogen atom and the third one (A- → A. + e-) would be related to the oxidation potential of the anion.
This acidity-derived approach was demonstrated by Frederick Bordwell at Northwestern University for a number of different families of compounds. The approach requires an empirical correction that can be applied for each family of compounds that account for differences in the ways the different thermodynamic parameters are measured. For example, a bond dissociation energy is usually treated as a gas phase problem, but acidity constants are measured in solution, so there are solvation energies that must be accounted for. These correction factors can be enormous. Nevertheless, if one bond strength within a family of compounds is already known, many others can be reliably calculated using a thermodynamic cycle and the appropriate correction.
Problem MK12.3.
Bordwell's team used this Hess' Law approach to calculate the bond strengths of phenol. (Frederick G. Bordwell and Jinpei Cheng, Substituent effects on the stabilities of phenoxyl radicals and the acidities of phenoxyl radical cations. J. Am. Chem. Soc. 1991, 113, 1736-1743). The oxidation potential of phenoxide ion, C6H5O- was determined to be 0.55 V (vs Fc+/Fc0), and they measured the acidity of phenol (pKa = 18). The reduction potential of a proton was known to be -0.40 V (vs Fc+/Fc0). They determined that at 298 K, BDE (kcal/mol) = 1.37pKa + 23.06Eoox(PhO-) + 23.06Eored(H+) + correction.
a) Show how they arrived at those factors, 1.37 and -23.06.
b) Calculate the BDE of the OH bond of phenol using Bordwell’s method, given the
correction = +46 kcal/mol.
This site was written by Chris P. Schaller, Ph.D., retired, College of Saint Benedict / Saint John's University (with contributions from other authors as noted). It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
Navigation: