Biochemistry Online: An Approach Based on Chemical Logic

 Homework
Problems - Literature Learning Module: Binding 1
Homework
Problems - Literature Learning Module: Binding 1
Assessment of Biochemistry/Molecular Biology (BMB) Foundational Concepts
Henry Jakubowski, Ph.D., Professor, Chemistry Department, College of Saint Benedict/Saint John's University
This page contains assessment/exam questions using data, figures, and graphs from research journals such as the Journal of Biological Chemistry which allow their use, or from journals such as from PLOS that are completely open access The papers and topics chosen were selected to assess student understanding of the American Society for Biochemistry and Molecular Biology (ASBMB) foundational concepts and learning objectives as well as MCAT2015 foundational concepts and objectives. These two sets of standards broadly overlap. Both ASBMB and the MCAT2015 strongly emphasis scientific inquiry and reasoning skills, which are perhaps best assessed by open-ended questions derived from the literature in which students must employ higher level Bloom skills of application and analysis.
These questions can also be used by students who seek more opportunities to practice interpreting research literature results. The ability to apply, analyze, and evaluate information and concepts are at the heart of scientific inquiry and reasoning skills which are central to the new ASBMB and MCAT2015 competency standards. The questions in this learning module are designed to assess these competencies using open-ended responses instead of multiple-choice questions. Answers can be found at the link at the bottom of this page.
Research Paper: Severe Acute Respiratory Syndrome Coronavirus 3C-like Proteinase N Terminus Is Indispensable for Proteolytic Activity but Not for Enzyme Dimerization: Biochemical and Thermodynamic Investigation in Conjunction with Molecular Dynamics Simulations. Shuai Chen, Lili Chen, Jinzhi Tan, Jing Chen, Li Du, Tao Sun, Jianhua Shen, Kaixian Chen, Hualiang Jiang, and Xu Shen. J. Biol. Chem., Vol. 280, Issue 1, 164-173, January 7, 2005
Background from paper
SARS coronavirus (SARS-CoV) has a replicase gene with a sequence motifs of papain-like proteinase and 3C-like proteinase (3CLpro). SARS 3CLpro is fully conserved among all released SARS coronavirus genome sequences. Because of its functional indispensability in the coronavirus life cycle, SARS 3CLpro (S3) has become an attractive target in discovering new anti-SARS agents. The crystal structures of 3CL proteinases from human coronavirus has been determined. The reported crystal structures of SARS 3CLpro and its complex with an inhibitor revealed substantially pH-dependent conformational changes and an unexpected model for inhibitor binding. It contains the following three domains: domains I and II (residues 1-184) have an antiparallel β-barrel structure forming a chymotrypsin-fold, the substrate-binding site is located in a cleft between these two domains; whereas residues 201-303 form a third compact α-helical domain (domain III) connecting to domain II by a long loop (residues 185-200).
As the crystal structures of 3CL proteinases in different coronaviruses give similar dimeric structures and nearly all side chains of 3CLpro involved in the formation of the dimer are conserved, it has been proposed that the dimer might be the biological functional form of 3CLpro. From the dimeric structures of coronavirus 3CLpros, it has been also predicted that domain III might play a role in substrate recognition and be responsible for positioning the N terminus of one protomer (monomer) to interact with the active site of the other protomer. The critical role of domain III in dimerization and enzymatic activity of SARS 3CLpro has been shown. In addition, Lai and co-workers reported that SARS 3CLpro exists as a monomer/dimer mixture at a relatively high protein concentration (approx. 4 mg/ml) and is exclusively monomer at a lower protein concentration (approx. 0.2 mg/ml) in solution, and only the dimer was the active form of the proteinase.
Based on the crystal structures and biochemical studies of several coronavirus 3CLpros, one hypothesis was proposed that the N-terminal residues 1-7 might play an important role in both dimerization and enzymatic activity of SARS 3CLpro. However, other studies showned that the N terminus was not necessary for the dimer formation of SARS 3CLpro, i.e. the N-terminal deleted SARS 3CLpro may possibly form a dimer with a new state different from the full-length proteinase. Biochemical and biophysical studies of the dimerization and proteolytic activities of the full-length (S3) and N-terminal residues 1-7 deleted SARS (S3-N) using CD spectroscopy, fluorescence resonance energy transfer (FRET), glutaraldehyde cross-linking SDS-PAGE, size-exclusion chromatography (SEC), and isothermal titration calorimeter (ITC) techniques are summarized below.
1. Interpret the following CD of the S3 and S3-N proteins. Why did they perform this experiment?
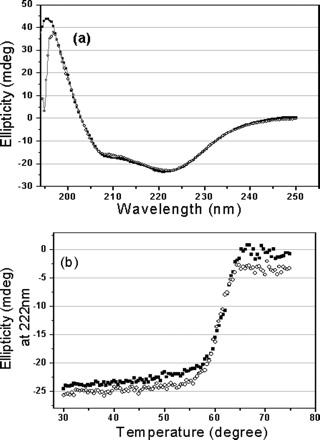
Fig 1. CD spectra of the full-length and N-terminal
deleted SARS 3CLpros. a, far-UV CD spectra of the
full-length (![]() ) and N-terminal
deleted (
) and N-terminal
deleted (![]() ) SARS 3CLpros at
25 �C. b, thermal induced unfolding monitored at 222 nm with a
temperature range from 30 to 75 oC for the full-length (
) SARS 3CLpros at
25 �C. b, thermal induced unfolding monitored at 222 nm with a
temperature range from 30 to 75 oC for the full-length (![]() )
and N-terminal deleted (
)
and N-terminal deleted (![]() ) SARS 3CLpros.
Protein concentrations used in CD experiments were 10 uM,
and all protein samples were prepared in 20 mM sodium
phosphate, pH 7.5, 100 mM NaCl.
) SARS 3CLpros.
Protein concentrations used in CD experiments were 10 uM,
and all protein samples were prepared in 20 mM sodium
phosphate, pH 7.5, 100 mM NaCl.
2. The enzymatic activity of the two proteins was studied using a novel fluorescent substrate (reactant), EDANS-VN_STLQSGLRK(Dabcyl)-M, which is totally hydrolyzed by papain-like protases. This is a short peptide with an amino terminus dansyl fluorophore and a C terminus Dabcyl group which does not fluoresce but can be excited by the excited state dansyl fluorophore ,which can transfer energy to the Dabcyl group in a non-radiative process, but only if it is very close in space to the dansyl group. A graph of enzyme activity is shown below.
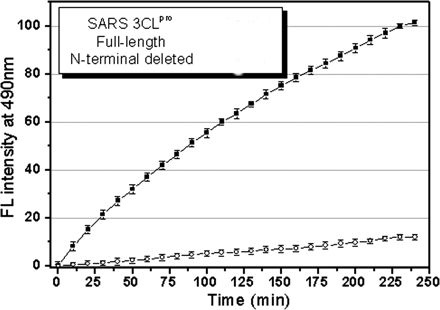
Fig. 2. Representative fluorescence profiles of
hydrolysis of the fluorogenic substrate by SARS 3CLpros. The
fluorogenic substrate at a concentration of 10 uM
was incubated with 1 uM full-length ()
or N-terminal deleted (![]() ) SARS 3CLpro
in 20 mM sodium phosphate, pH 7.5, 100 mM
NaCl, 5 mM DTT, 1 mM EDTA, at 25
o C. Increase of emission fluorescence intensity at 490 nm wavelength was
recorded at 10-min intervals,
) SARS 3CLpro
in 20 mM sodium phosphate, pH 7.5, 100 mM
NaCl, 5 mM DTT, 1 mM EDTA, at 25
o C. Increase of emission fluorescence intensity at 490 nm wavelength was
recorded at 10-min intervals, ![]() EX
= 340 nm. The emission spectrum was recorded for more than 4 h. The initial
reaction velocity (
EX
= 340 nm. The emission spectrum was recorded for more than 4 h. The initial
reaction velocity (![]() 0) was
determined from the linear portion of the progress curve, which corresponded to
between 2 and 10% hydrolysis of the substrate. The activity of the full-length
SARS 3CLpro was taken as 100%.
0) was
determined from the linear portion of the progress curve, which corresponded to
between 2 and 10% hydrolysis of the substrate. The activity of the full-length
SARS 3CLpro was taken as 100%.
a. Give a molecular explanation of why the fluorescence increases on S3 cleavage of the substrate
b. What conclusions can you draw concerning the different activity of S3 and S3-N towards the substrate?
3. One way to determine if a protein can dimerize is to introduce covalent bonds between adjacent proteins subunits in potential dimer or multimer. One such cross-linking reagent is glutaraldehye, which is shown below.

a. Draw a mechanism to show how it could react with Lys residues on two adjacent proteins to crosslink them.
SDS-PAGE gels were run for both S3 and S3-N, and are shown below.
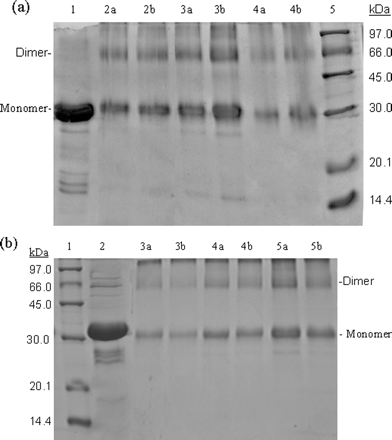
Fig 3. SDS-PAGE (10% acrylamide) profiles of
glutaraldehyde cross-linked SARS 3CLpros.
a, cross-linking
analyses of the full-length SARS 3CLpro. Lane 1, untreated 3CLpro
(7 mg/ml); lane 2a, 3CLpro (2 mg/ml) cross-linked by 0.01%
glutaraldehyde; lane 2b, 3CLpro (2 mg/ml) cross-linked by 0.1%
glutaraldehyde; lanes 3a and 3b, 3CLpro (7 mg/ml)
cross-linked by 0.01 and 0.1% glutaraldehyde, respectively; lanes 4a and
4b, 3CLpro (1 mg/ml), 0.01 and 0.1% glutaraldehyde; lane 5,
molecular weight SDS calibration kit protein standards are as follows:
phosphorylase b (97.0 kDa), albumin (66.0 kDa), ovalbumin (45.0 kDa),
carbonic anhydrase (30.0 kDa), trypsin inhibitor (20.1 kDa),
![]() -lactalbumin (14.4 kDa).
b,
cross-linking analyses of the N-terminal deleted SARS 3CLpro. Lane
1, protein standards; lane 2 untreated 3CLpro (7 mg/ml);
lanes 3a and 3b, 3CLpro (1 mg/ml), 0.01 and 0.1%
glutaraldehyde, respectively; lanes 4a and 4b, 3CLpro
(2 mg/ml), 0.01 and 0.1% glutaraldehyde; lanes 5a and 5b, 3CLpro
(7 mg/ml), 0.01 and 0.1% glutaraldehyde.
-lactalbumin (14.4 kDa).
b,
cross-linking analyses of the N-terminal deleted SARS 3CLpro. Lane
1, protein standards; lane 2 untreated 3CLpro (7 mg/ml);
lanes 3a and 3b, 3CLpro (1 mg/ml), 0.01 and 0.1%
glutaraldehyde, respectively; lanes 4a and 4b, 3CLpro
(2 mg/ml), 0.01 and 0.1% glutaraldehyde; lanes 5a and 5b, 3CLpro
(7 mg/ml), 0.01 and 0.1% glutaraldehyde.
b. What conclusions can you draw concerning the ability for the two proteins to form dimers??
4. To further study the solution behavior of the protein, they were subjected to gel filtration (also called size exclusion chromatgraphy) on Superdex 75. Results are shown in Figure 4 below.
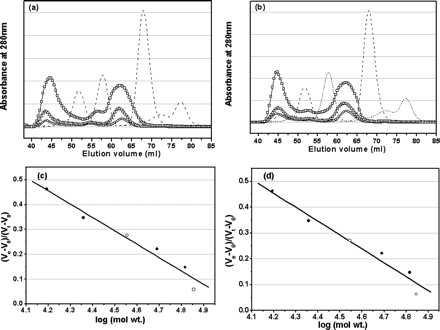
Fig 4. Analyses of monomer-dimer equilibria of SARS 3CLpros
by SEC. a, elution profiles of the full-length SARS 3CLpro
at neutral pH (7.5) and concentrations of 1 (![]() ),
2 mg/ml (
),
2 mg/ml (![]() ), and 7 mg/ml (
), and 7 mg/ml (![]() ).
Elution profiles of four marker proteins, ribonuclease A (15.6 kDa),
chymotrypsinogen A (22.8 kDa), ovalbumin (48.9 kDa), and albumin (65.4 kDa), are
also shown as dashed lines. The dimer/monomer ratios for the
concentrations of 1, 2, and 7 mg/ml are 0.68, 0.71, and 0.84, respectively.
b,
elution profiles of the N-terminal deleted SARS 3CLpro at neutral pH
(7.5) and concentrations of 1 (
).
Elution profiles of four marker proteins, ribonuclease A (15.6 kDa),
chymotrypsinogen A (22.8 kDa), ovalbumin (48.9 kDa), and albumin (65.4 kDa), are
also shown as dashed lines. The dimer/monomer ratios for the
concentrations of 1, 2, and 7 mg/ml are 0.68, 0.71, and 0.84, respectively.
b,
elution profiles of the N-terminal deleted SARS 3CLpro at neutral pH
(7.5) and concentrations of 1 (![]() ), 2 (
), 2 (![]() ),
and 7 mg/ml (
),
and 7 mg/ml (![]() ). Elution profiles of
marker proteins are shown as dashed lines. The dimer/monomer ratios for
the concentrations of 1, 2, and 7 mg/ml are 0.65, 0.66, and 0.80, respectively.
In the SEC-FPLC experiment, each protein sample was loaded to a HiLoad 16/60
Superdex 75 prep grade column pre-equilibrated with the buffer and then eluted
at a flow rate of 1 ml/min with a detection of absorbance at 280 nm. The buffer
used was 20 mM sodium phosphate, pH 7.5, 100 mM
NaCl, 5 mM DTT, 1 mM EDTA.
Standard calibration curves were used to determine the molecular masses of
monomeric and dimeric form of the full-length (c) and N-terminal deleted
(d) SARS 3CLpros. Elution volumes of the full-length and
N-terminal deleted proteinases are indicated by open circles (
). Elution profiles of
marker proteins are shown as dashed lines. The dimer/monomer ratios for
the concentrations of 1, 2, and 7 mg/ml are 0.65, 0.66, and 0.80, respectively.
In the SEC-FPLC experiment, each protein sample was loaded to a HiLoad 16/60
Superdex 75 prep grade column pre-equilibrated with the buffer and then eluted
at a flow rate of 1 ml/min with a detection of absorbance at 280 nm. The buffer
used was 20 mM sodium phosphate, pH 7.5, 100 mM
NaCl, 5 mM DTT, 1 mM EDTA.
Standard calibration curves were used to determine the molecular masses of
monomeric and dimeric form of the full-length (c) and N-terminal deleted
(d) SARS 3CLpros. Elution volumes of the full-length and
N-terminal deleted proteinases are indicated by open circles (![]() )
and for marker proteins by filled circles. Ve and
V0 are peak elution volume and column void volume,
respectively, and Vt is the total bed volume.
)
and for marker proteins by filled circles. Ve and
V0 are peak elution volume and column void volume,
respectively, and Vt is the total bed volume.
a. Why did they vary the concentration of each protein.
b. What conclusions can you draw concerning the ability for the two proteins to form dimers?
c. The integrated areas of the peaks give dimer/monomer ratios of 0.68, 0.71 and 0.84 for protein concentrations of 1, 2, and 7 mg/ml. Write the chemical and mathematical equilibrium expression for the process and using these, explain if the variation of the dimer/monomer ratio is expected.
5. The monomer/dimer interaction was then studied by isothermal titration calorimetry. In the experiments, concentrated protein (around 500 - 600 μM or 16-17 mg/ml) was injected into a buffer solution and enthalpy changes measured. Results are shown below. Note that all injections of the protein give + enthalpy values indicated the process is endothermic.
5. 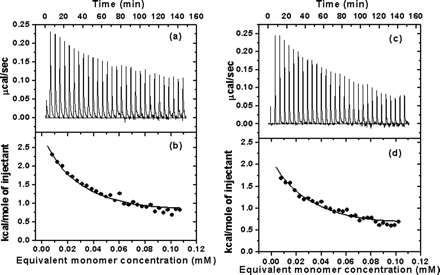
Fig 5. Typical calorimetric dilution data measured by ITC. a, the calorimetric dilution profile for full-length SARS 3CLpro dimers. Consecutive 10 ul volumes of 572.5 uM 3CLpro (approx 17 mg/ml) were diluted into 1.43 ml of 20 mM sodium phosphate, pH 7.5, 100 mM NaCl, 1 mM EDTA, at 25 oC (except an initial injectant of a 3-�l volume). Note that the protein was dissolved in the same buffer solution. b, integrated dilution heat effects of full-length SARS 3CLpro, with theoretical fits by a simple monomer-dimer model to yield a Kd of uM and a dimerization enthalpy change (ΔHodimer) of -8.283 + 0.103 kcal/mol. c, the calorimetric dilution profile for N-terminal deleted SARS 3CLpro. Endothermic responses for sequential 10 ul injections of 544.6 uM 3CLpro into buffer were measured under the same conditions, except a 3 ul initial injection. d, integrated dilution heat effects of N-terminal deleted SARS 3CLpro , with theoretical fits to yield a Kd of 262 + 15 uM and a ΔHodimerization of -6.893 + 0.25 kcal/mol.
a. Write a chemical equation that clearly shows what process occurs when the first injection of protein S3 is made into the calorimeter cell. Calculate the initial concentration of S3 at the instant (t=0) it is injected into the calorimeter cell. What is the state of the diluted protein at t=0 and at the end of the time increment indicated by return of the enthalpy curve to the base line value AFTER THE FIRST INJECTION (i.e. at the end of the spike in the enthalpy for the first 10 ul protein injection).?
b. Why does the enthalpy change becomes smaller and smaller (as shown in Fig 5b) as more concentrated protein is added to the calorimetry cells.
c. Calculate ΔG0 and ΔS0 for the dimerization equilibrium for S3.
d. What conclusions can you draw concerning the ability for the two proteins to form dimers?
6. Molecular dynamics simulations were made from the pdb file for S3 (in which the unit cell contained a dimer) and models based on it for S3-N. Modeling shows that S3-N forms a dimer but with a reduced substrate binding pocket. To confirm this, a small peptide substrate, TSAVLQ, was synthesized. This represented the N terminal sequence of the best reported peptide substrate for the enzyme. The binding affinity of the peptide for S3 and S3-N was determine by surface plasmon resonance biosensors. S3 or S3-N were immobilized to cells for sensor chip from Biacore. Buffer was continuously flowed over the derivitized chip. The peptide was diluted into the buffer solution and injected in increasing concentrations from 20-500 μM. Binding responses were recorded continuously in resonance units (RU) as sensorgrams. The results are shown below.
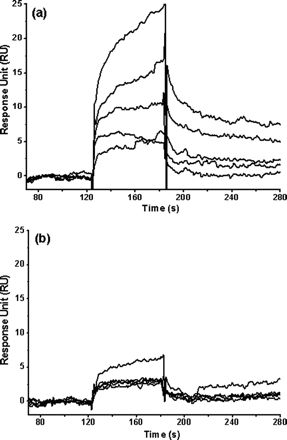
Fig 7. Binding affinity of a 6-amino acid peptide (TSAVLQ) to the full-length and N-terminal deleted SARS 3CLpros determined by SPR. Real time binding affinity measurements of the peptide to the full-length SARS 3CLpro (a) and the N-terminal deleted proteinase (b) using Biacore 3000. Representative sensorgrams were obtained from injections of the peptide at concentrations of 500, 250, 100, 50, and 20 �M (curves from top to bottom). The peptides were injected for 60s (from 120-180 seconds), and dissociation was monitored for more than 120s (from 180 to 300 seconds).
a. Write the chemical equilibrium expression for the protein and peptide interaction showing appropriate rate constants. What constant (Kd, kforward, or kreverse can be determined from data in the range 120-180s? 180-300s?
b. From kforward and kreverse, how could you determine Kd?
c. Compare and interpret the two graphs. Are the data consistent with the other experiments?
Answers: Literature Learning Module: Binding 1
Navigation
Return to Biochemistry Online Table of Contents

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.