Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
C: MODEL BINDING SYSTEMS
BIOCHEMISTRY - DR. JAKUBOWSKI
3/29/16
|
Learning Goals/Objectives for Chapter 5C: After class and this reading, students will be able to
|
C7. Binding to Intrinsically Disorder Protein and MORFs
As described above, binding of a protein to a ligand (including another protein) could occur by a lock and key mechanism, possibly through a conformational selection process, or through an induced fit when an initial binding event is followed by a conformation rearrangement to form a more tightly bound complex. But how does binding to completely intrinsically disorder protein (which has been documented) occur? These cases are quite removed from those envisioned in simple induced fit mechanisms. To accommodate binding to IDPs, a new idea has emerged, that of Molecular Recognition Features (MoRFs).
MoRFs are typically contiguous but disordered sections of a protein that first encounter a binding partner (a protein for example). Using protein complexes in the Protein Data Bank, Mohan et al conducted a structural study of MoRFs by selecting short regions (less than 70 amino acids) from mostly disordered proteins that were bound to proteins of greater than 100 amino acids. They chose a sequence size of 70 amino acids and smaller since they would be most likely to display conformational flexibility before binding to a target. 2512 proteins fit their criteria. For comparison, they created a similar database of ordered monomeric proteins. The analysis showed that after they encounter a binding surface on another protein, the MoRF would adopt or "morp" into several types of new conformations, including alpha-helices (a-MoRFs), beta-strands (b-MoRFs), irregular strands (i-MoRFs) and combined secondary structure (complex-MoRFs), as shown in the figure below.
Figure: Types of Molecular Recognition Features in Intrinsically Disordered Proteins
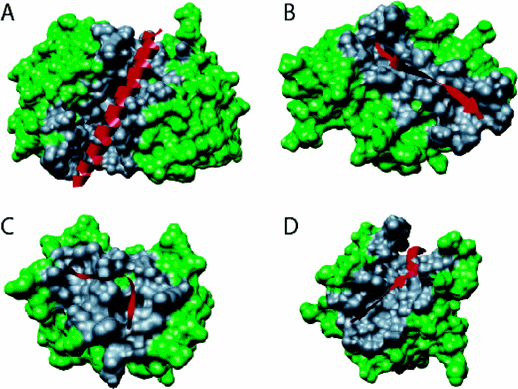
(A) α-MoRF, Proteinase Inhibitor IA3, bound to Proteinase A (PDB entry 1DP5). (B) A β-MoRF, viral protein pVIc, bound to Human Adenovirus 2 Proteinase (PDB entry 1AVP). (C) An ι-MoRF, Amphiphysin, bound to α-adaptin C (PDB entry 1KY7). (D) A complex-MoRF, β-amyloid precursor protein (βAPP), bound to the PTB domain of the neuron specific protein X11 (PDB entry 1X11). Partner interfaces (gray surface) are also indicated. Vacic, V. et al. Journal of Proteome Research 6, 2351 (2007). Permission from Copyright Clearance Center's Rightslink /American Chemical Society
Vacic et al have further characterized the binding surfaces between MoRFs and their binding partners using structural data from PDB files. Interfaces were studied by determining the differences in accessible surface area between MoRFs and their binding partners, and the protein in unbound states. These were compared to ordered protein complexes, including homodimers and antibody-protein antigen interactions that were not characterized by disordered interactions. Their findings are summarized below.
-
MoRF interfaces have more hydrophobic groups and fewer polar groups compared to the surface of monomers. This is true even as the overall amino acid composition of intrinsically disordered protein shows them to be enriched in polar amino acids, which leads them to adopt a variety of unfixed solution conformations.
-
a-MoRFs have few prolines, which is expected as prolines are helix breakers.
-
Methionine is enriched in both MoRFs and in their binding partner interface. Methionine is unbranched, flexibile, and contains sulfur, which is large and polarizable, making it an ideal side chain to be involved in London forces in a hydrophobic environment.
-
Even though MoRFs have few residues, their binding interfaces were of similar or larger size than other protein binding interfaces, a result which also applies to IDPs as a whole. MoRFs interfaces also have a larger solvent-exposed surface area, similar to IDPs. This is consistent with the notion MoRFs are disordered before binding and that a defined structure is not possible with little buried surface area.
-
As MoRFs have significant nonpolar character within a IDP that is highly enriched in polar amino acids, MoRFs should be highly predictable by search algorithms.
Navigation
Return to Chapter 5C: Model Binding Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5C: Model Binding Systems

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.