Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
C: MODEL BINDING SYSTEMS
BIOCHEMISTRY - DR. JAKUBOWSKI
3/29/16
|
Learning Goals/Objectives for Chapter 5C: After class and this reading, students will be able to
|
C9. Binding and Linked Equilibria
Hemoglobin is an amazing molecule. Unlike myoglobin, in displays cooperative binding of dioxygen. In addition, not only does it bind dioxygen (at the heme Fe), it also binds protons and carbon dioxide at other, or allosteric sites. Binding of these ligands influences the binding of dioxygen, as seen from the right ward shift of the oxygen binding curves at lower pH's, such as would be encountered in respiring tissues. The linkage of the binding of dioxygen, and protons or carbon dioxide can be visualized easily with simple thermodynamics which you learned in general chemistry. Consider the equilibria shown below.
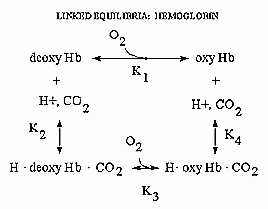
It should be obvious from these equations than K1 K4 = K2 K3 , where Ki are the dissociation constants for the various equilibrium. Remember than ΔG is a state function and independent of path. Hence ΔGo1 + ΔGo4 = ΔGo2 + ΔGo3 , where ΔGoi = -RTln(1/Kdi). From the oxygen binding graphs, it is also clear that K1 < K3. Therefore, it is clear that K2 < K4. This shows that protons binds to deoxyHb more tightly than to oxyHb. Hence, the presence of higher [H+] drives the dissociation of O2 from oxyHb. As described in the book, H+ bind to side chains which have an increased affinities for H+ in the deoxyHb state. The conformation change in Hb alters the effective environment of some side chains (specifically His β146, His α122, and the α amino group of the α chain), increasing their pKa values The protonated HisH+ then makes a salt bridge to Asp β 94 in the FG corner of the same β chain, forming a salt link which stabilizes the deoxyHb or taut state.
Figure: A review of thermodynamic cycles - linked equilibria - other examples
Navigation
Return to Chapter 5C: Model Binding Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5C: Model Binding Systems

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.