Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
D: BINDING AND THE
CONTROL OF GENE
TRANSCRIPTION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/30/16
|
Learning Goals/Objectives for Chapter 5D: After class and this reading, students will be able to
|
D13. Control of Gene Expression by RNA
What accounts for the increased complexity of organisms like humans? As was discussed in the DNA chapter, it is not the number of chromosomes or even the number of possible genes in an organism. One big difference between bacterial and human cells, for example, is the percentage of DNA coding for proteins. In bacteria, most of the DNA codes for proteins, but in human eukaryotic cells, most of the DNA (up to 98%) is "junk" in that it does not code for proteins. The DNA consists of intervening sequences within DNA coding for a given protein, and sequences between genes. Up to 98 % of the RNA transcribed in human cells is derived from this "junk" DNA. What function does this RNA serve? New evidence shows that this transcribed RNA binds to other RNA molecules like mRNA (to inhibit its translation), to DNA (to control gene transcription) or to proteins (to alter gene transcription as well). These process are called RNA interference (RNAi).
An understanding of RNAi really began in 1998 with the study of RNAi in round worms (described below) by Fire and Mellon, who were awarded the Nobel Prize in Physiology and Medicine in 2006. The Nobel Foundation stated that "the discovery of RNAi has already had an immense impact on biomedical research and will most likely lead to novel medical applications in the future".
The terminology used to describe the RNA species involved in RNAi is often confusing (especially to a protein chemist). In part it depends on the sources of the RNA. The RNA can derive from the cells own endogenous RNA (transcribed from genes of the cell) or by exogenous RNA entering the cell from the outside. This can happen by infection by an RNA virus, a DNA virus (which forms RNA in the cell after transcription), by plasmids containing DNA that will be transcribed to RNA in the cell, or by synthetic RNA. These RNAs generally inhibit translation of mRNA for a given protein and are described below in more detail.
RNAi from enodgenous RNA
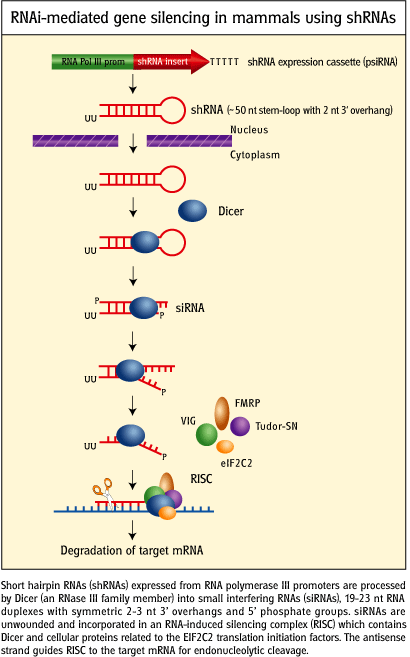
- microRNAs (miRNAs) are formed from endogenous RNA arising from transcription of certain stretches of nuclear DNA. From these regions, the transcribed RNA can base pair on itself to form a double stranded stem-loop hairpin structure. The initial transcript is call primary miRNA (or pri-miRNA). The pri-miRNA are transcribed by RNA polymerase III and are post-transcriptionally process to contain a modified 5' nucleotide and a 3' poly A tail The pri-miRNA folds to form regions stem-loop structures where the ds-stem is held together by intracchain H bonds. This stem-loop motif is recognized by an enzyme called Drosha, which cleaves it to form a separated stem:loop structure of about 70 nucleotide (NT) size (often called a pre-miRNA) with nonoverlapping ends. The stem:loop structures also can be called short hairpin RNA (shRNA). The dsRNA of the stem loop is recognized by an enzyme called dicer which removes the loop leaving a dsRNA of about 20-25 NTs in length. The short dsRNAs can then bind to a protein complex called RISC (RNA-induced silencing complex), which promotes unraveling of the dsRNA. A complex of RISC and one of the short RNA strands can bind to complementary stretches in mRNA for a specific gene (viral or host) and inhibit translation of the mRNA into protein. Inhibition occurs when RISC complex cleaves the RISC-siRNA- mRNA complex. (i.e one of the components in the RISC complex is an RNA nuclease.) This mechanism might be linked to defense mechanisms of virally-infected cells by inhibition of viral mRNA translation. If the complementarity of the mRNA and miRNA is less than ideal (which it usually is for miRNA), then binding of miRNA to the mRNA may only attenuate the translation of the mRNA. miRNA are often complementary to the 3' untranslated region (UTR) of the mRNA. It is estimated that the human genome has over 1000 miRNA which are used to regulate gene transcription (through attenuation of protein synthesis.
RNAi from exogenous RNA (or DNA)
- short interfering RNAs (siRNAs): These are synthesized in the lab and can be used to interfere with translation when added to cells. They are made to be perfectly complementary to a mRNA target. Often three different siRNA are used to target different regions of the target mRNA. As these are synthetic, their main function is in experimental studies to probe the effect of knocking down the expression of a gene by temporarily decreasing translation from a specific mRNA. These knockdowns are temporary as the siRNA degrade in the cell and are not replicated. Modified bases can be used in the synthesis of the siRNA to increase their half-life in the cell. It is difficult to get siRNAs into the cell (in a process called transfection) to produce these alterations in gene expression. Special transfection reagents have been developed to facilitate that process.
- short hairpin RNA (shRNA): instead of adding siRNA directly, a plasmid DNA containing a gene that when transcribed in the cell can form a shRNA can be added to the cell. Uptake of plasmids containing human genes by prokaryotes (in a process called transformation) is the most common way to make human proteins in bacterial cells. The plasmids can be replicated so the transformation of cells that take up the plasmid is passed on to progeny cells. Likewise eukaryotic cells can be transfected (unfortunately the word transformation is not used for eukaryotic cells) by plasmids with genes encoding shRNA. Knockdowns produced by this technique would be much longer lived that those produced by the addition of synthetic siRNAs.
- lentiviral transfection with shRNAs: Lentiviruses (like HIV) contain a ssRNA genome. It can be modified to contain a gene for a shRNA. Once in the cells, the lentilviral RNA can be converted to DNA which is integrated covalently into the host cell genome. When transcribed, shRNA is produced which when processed byh diceer produces RNA interference in the cell. This transfection of cells is essentially permanent, allowing knockdowns of protein expression in progeny cells.
RNAi pathways probably evolved from or with pathways for cellular resistance to viruses. Viruses often produced dsRNA during their life cycle. Some viruses like the HIV virus have a ssRNA genome. One method of host defense against viral infection is fomation of short interfering RNAs that could inhibit transcription of viral proteins from viral mRNA.
![]() Jmol:
dsRNA (still figuring out to convert to JSMol)
Jmol:
dsRNA (still figuring out to convert to JSMol)
One of the first amazing demonstrations of the power of RNAi to module gene expression at the translational level was done in the nematode worm, C. elegans. This organism has about 20,000 genes which code for proteins. Kamathk et. al. fed these worms E. Coli transformed with plasmid DNA designed to produced dsRNA upon transcription, one strand of which was complementary to mRNA sequences in the worm. Plasmids containing almost 17,000 different dsRNA encoding genes were constructed and used to knockout gene expression by forming dsRNA complexes of the mRNA with the RNAi. Phenotypic changes in the organism were studied. About 1700 of the dsRNA experiments led to observable (phenotypical) changes in the organism. Genes whose inactivation was lethal (and hence were essential for survival) were generally those that had counterparts in all other organism, while those associated with nonlethal changes were more likely to be homologous to genes in higher organisms and more recently evolved. They also selectively looked at which genes influenced lipid metabolism by incorporating a fluorescent tag which bound to lipid deposits in the organism. Around 300 genes were found to influence fluorescence and hence regulate fat deposition in the organism.
Figure: RNA Interferene: Antisense and Silencing
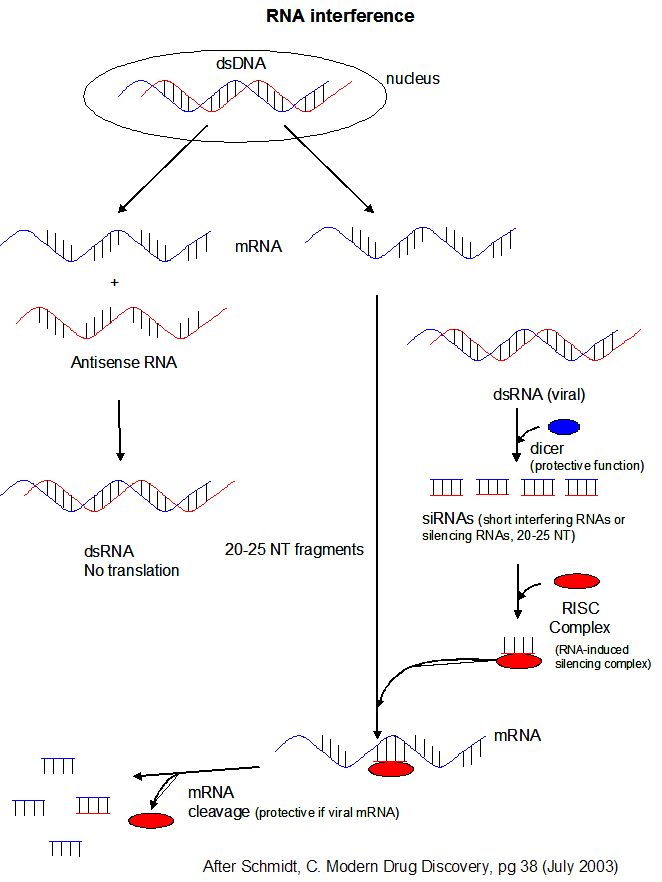
-
 Animation:
RNAi Web
Animation:
RNAi Web
RNAi is the basis of an new emerging industry. Many companies offer kits and free software that make RNAi studies simple. Invitrogen is one such company.
Figure: RNAi-mediated gene silencing in mammals using short haripin
RNA genes.
credit:
http://www.invivogen.com/sscat.php?ID=14
![]() Jmol: Updated
Dicer
Jmol14 (Java) |
JSMol (HTML5) Dicer
Jmol: Updated
Dicer
Jmol14 (Java) |
JSMol (HTML5) Dicer
Two groups have deleted miRNA-155 and looked at effects on immune cells in mice. Immune cell function in B, T, and dendritic cells was affected, leading to animal death when exposed to salmonella after they were immunized. Animals in sterile environments showed no effect. In contrast to knockouts of protein-coding genes, these knockouts affect transcription of multiple genes. Knockout of miR-208 caused heart problems in mice placed in a stressful environment. These experiments indicated that some genetic diseases might arise from mutations in non-protein coding regions of the genome.
Recently, a new mechanism in control of gene expression has been offered which involves regulation of translation of a mRNA. mRNA must have a sequence, the Shine-Delgarno sequence, which allows it to bind to ribosomes. If a ligand binds to this site, mRNA could not bind to the ribosome and translation would be inhibited. Such is the case in the mRNA encoding proteins involved in the transport and synthesis of vitamins B1 (thiamine) and B12 (adenosyl cobalamin). Thiamin and thiamine pyrophosphate were shown to bind to the leader sequence of an E. Coli mRNA involved in thiamine biosynthesis and inhibit the translation of the mRNA. This allosteric mechanism for inhibition makes physiological sense since the presence of high levels of cellular B1 would obviate the need for its synthesis or transport.
Navigation
Return to Chapter 5D: Binding and the Control of Gene Transcription
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5D: Binding and the Control of Gene Transcription

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.