Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
D: BINDING AND THE
CONTROL OF GENE
TRANSCRIPTION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/30/16
|
Learning Goals/Objectives for Chapter 5D: After class and this reading, students will be able to
|
D14. Recognition of viral and bacterial DNA by the immune system
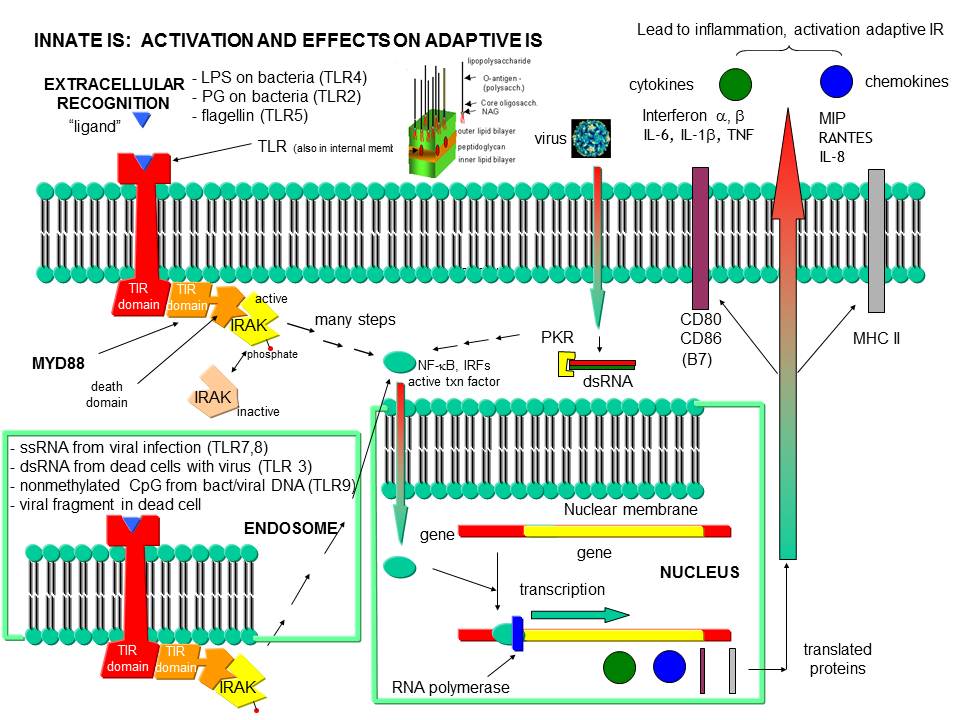
Before leaving the topic of RNA/proteins interactions, consider how a self cell would detect viruses and bacteria. It would be beneficial to the organism if the immune system could recognize and response to many types of bacteria, viruses, fungi, or protozoa by binding to common target on them. For example, it would be desirable to have a single cell type, such as a scavenging macrophage, have a recognition system that would recognize a common molecular pattern such as LPS found on gram negative bacteria. The part of the immune system that has this capability is called the innate immune system. The cells of the innate system (dendritic cells, macrophages, eosinophils, etc) have receptors (Toll-like Receptors 1-10 or TLRs) that recognize the common pathogen associated molecular patterns - PAMPs (sometimes called MAMP - microbe associated patterns), which leads to binding, engulfment, signal transduction, maturation (differentiation), antigen presentation, and cytokine/chemokine release from these cells. Take for example dendritic cells, which reside in the peripheral tissues and act as sentinels. They can bind PAMPs which include:
- CHO/Lipids on bacteria surface (LPS)
- mannose (CHO found in abundance on bacteria, yeast
- dsRNA (from viruses)
- nonmethylated CpG motiffs in bacterial DNA
TLR receptors are expressed on the cell surface for recognition of external PAMPS on foreign invaders. However, since bacterial and viral can be engulfed, it would be optimal to have intracellular recognition of viral and bacterial nucleic acids as well. These are recognized by intracellular TLRs in the cell after the they been taken up into the cells by endocytosis. The figure below shows how viral and bacterial nucleic acids found in endosomal vesicles, can be bound by endosomal membrane TLRs. A Jmol model of a recent structure of TLR3 and dsRNA is shown below.
Figure: Endosomal TLR3 Interaction with foreign RNA and DNA
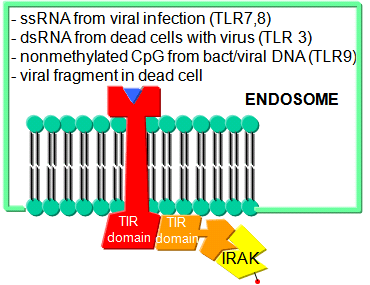
![]() Jmol: Updated
TLR3:dsRNA complex
Jmol14 (Java) |
JSMol (HTML5) Needs updating
Jmol: Updated
TLR3:dsRNA complex
Jmol14 (Java) |
JSMol (HTML5) Needs updating
Inflammation can also arise when normal tissue is damaged due to injury, which exposes molecules usually located inside of the cell to the immune system. Such molecules include high mobility group proteins (associated with chromatin), proteoglycans and nucleic acids. These are referred to as damaged associated molecular pattern (DAMP) molecules. Intracellular proteins exists normally in a reducing environment so when they are exposed to the oxidizing conditions of the extracellular milieu, covalent and conformational changes may ensue that
Figure: Recognition of PAMPs by TLRs
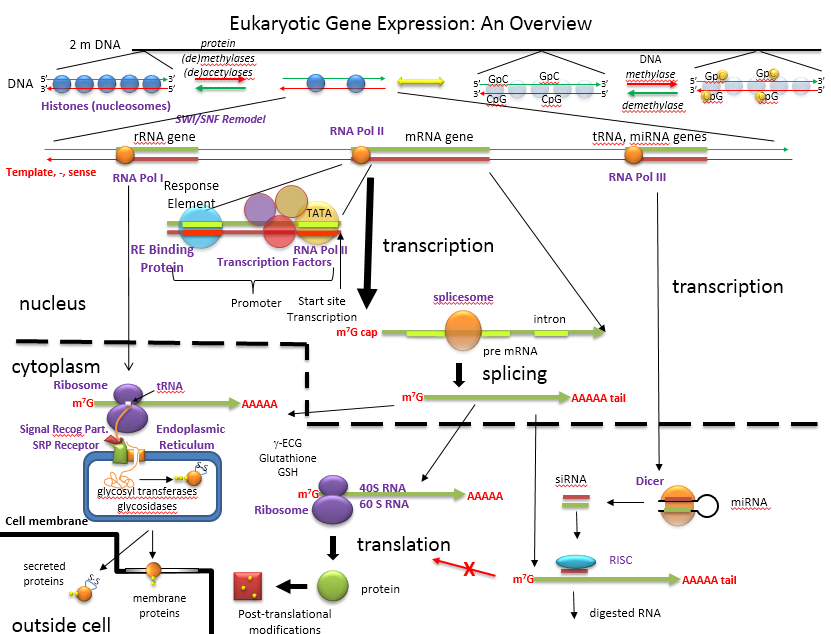
The figure below shows an overview of how the genetic information is decoded into proteins and how those processes are controlled.
An Overview of the Regulation of Gene Expression in Eukaroytes
Navigation
Return to Chapter 5D: Binding and the Control of Gene Transcription
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5D: Binding and the Control of Gene Transcription

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.