Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 5 - BINDING
E: MODERN METHODS IN DRUG DEVELOPMENT
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Updated: 03/31/16
|
Learning Goals/Objectives for Chapter 5E: After class and this reading, students will be able to
|
E5. DNA Binding Drugs
Most drugs in use today bind to specific proteins and interfere with the activity of the protein. Some antibiotics do bind DNA by intercholating nonspecifically between base pairs and interfering with replication and transcription. An alternative new approach is to design drugs that bind to specific sequences of DNA and either promote or inhibit the transcription of key genes. β-hydroxybutyrate is an experimental drug now being used to treat sickle-cell anemia. Somehow it increases the transcription of fetal hemoglobin genes, which form proteins which do not aggregate as the sickle cell hemoglobin gene product does. These approach has been called chemical genetics. Stuart Schreiber at Harvard has been a leader in the this field.
-
 Article
on Schreiber's work
Article
on Schreiber's work -
 Schreiber's
company, Infinity Pharmaceuticals
Schreiber's
company, Infinity Pharmaceuticals
Another way to inhibitor specific gene transcription is to bind a small single-stranded DNA to the dsDNA to form a triple helix. The single stranded DNA molecule can bind to exposed H bond donors and acceptors of bases not involved in Watson-Crick H-bonding interactions between complementary bases in the ds DNA.
Figure: Triple helix
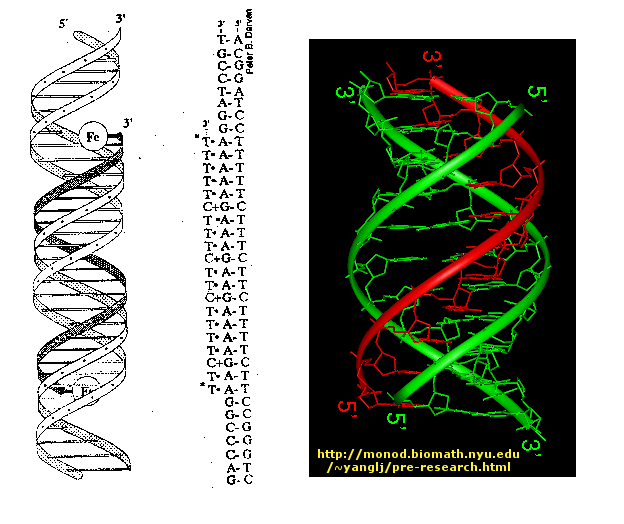
The single strand binds to such donors that are accessible in the major grove of dsDNA.
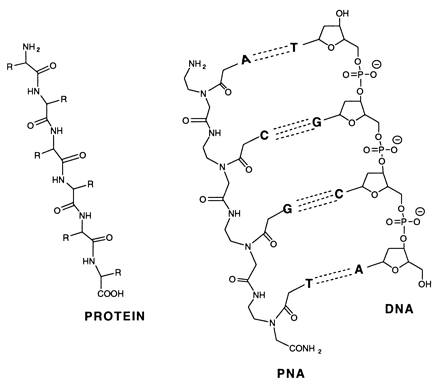
Peptide-nucleic acid analogs, as shown below, are promising new drugs. They are less charged than nucleic acids (so can more easily cross a membrane) and are resistance to cleavage by proteases and nucleases. One could image them binding to specific nucleotide sequences and inhibiting processes like DNA replication and transcription.
Transcription factors that are generally activated by genetic events or upstream signaling pathways are key regulators of cell state. Due to the extensive protein-protein interfaces and general absence of hydrophobic pockets that might inhibit protein:protein interactions required for transcriptional regulation, it has been difficult to design drugs that bind to transcription factors and modulate their activity. Moellering et al. report a successful development of a direct-acting antagonist of an oncogenic transcription factor, NOTCH1. This antagonist consists of cell-permeable stabilized α-helical peptides, SAHMs that was "stapled" into a stable helix through addition of two unnatural alkenyl amino acids which through ring closure sterically restrained the peptide in an alpha helix. The helix mimicked one protein:protein interface region in the ternary complex of DNA:NOTCH1:MAML1 (which is a coactivator protein). The peptides antagonized on NOTCH signaling and cell proliferation in T-cell acute lymphoblastic leukemia cells (T-ALL).
Navigation
Return to 5E. Modern Methods in Drug Development Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 5E: Modern Methods in Drug Development

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.