Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8: OXIDATIVE-PHOSPHORYLATION
A: THE CHEMISTRY OF DIOXYGEN
BIOCHEMISTRY - DR. JAKUBOWSKI
04/14/16
|
Learning Goals/Objectives for Chapter 8A: After class and this reading, students will be able to
|
A8. Just Say NO - The Chemistry of Nitric Oxide
In the last decade, the role of another gaseous free radical, nitric oxide (.NO) has become apparent. This molecule is synthesized biologically through the action of an inducible heme enzyme, nitric oxide synthase, which forms NO by the oxidation by dioxygen of the guanidino group of Arg, which gets converted to citrulline. .NO is soluble and can diffuse through cell membranes into the cytoplasm, where it has a myriad of effects in signal transduction pathways. However, it can also be metabolized to form reactive nitrogen intermediates (much as with dioxygen) which can be deleterious to the body, if they damage native biomolecules, or advantageous, when they are used by immune cells like macrophages in the destruction of engulfed bacteria.
A molecular orbital diagram of .NO shows it to have a bond order of 2.5 and one unpaired electron in a π2p* antibonding orbital (hence the notation .NO).
Figure: Molecular Orbital Diagram of NO
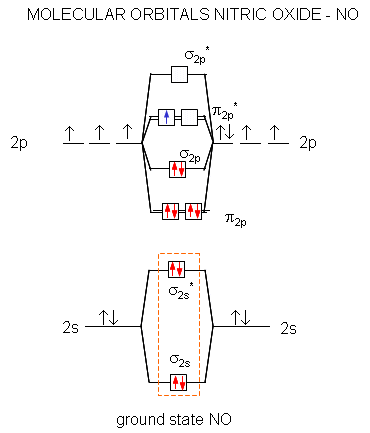
Here are some of the relevant reactions of NO and its reactive nitrogen intermediates (RNI):
-
The high energy electron π2p*can leave on oxidation of NO to form the nitrosyl ion, NO+. The bond order in NO+ is three suggesting strong interactions between the N and O atoms.
-
.NO (oxidation number of N of +2) is thermodynamically unstable and can disproportionate or dismutate (like superoxide) in a self redox reaction to form nitrous oxide (N2O) and nitrogen dioxide (.NO2), with oxidation number for N of +1 and +4, respectively.
-
In acidic condition .NO can be oxidized to nitrite (NO2-) which can be protonated to form nitrous acid (HNO2).
-
Nitrous acid can disproportionate or dismutate to form .NO and the radical .NO2 which can be protonated to ONOOH with a pKa of 6.5.
-
.NO can react with superoxide (.O2-) faster than any other molecule to form peroxynitrite, ONOO- . .NO and superoxide (.O2-) are both produced by cells during infllammation, leading to increased levels of peroxynitrite.
-
The O-OH bond in protonated peroxynitrite (ONO-OH) can be cleaved either heterolytically to produce NO3- and H+ (70%) or homolytically to produce the radical .NO2 and the highly reactive .OH free radical (30%).
- The deprotonated form of peroxynitrite (ONOO-) can oxidize organic molecules, especially thiols and CO2. When it reacts with CO2, it can form either NO3- and CO2 (65%) or CO3.- and .NO2 (35%). These latter products can oxidize organic molecules in one electron steps. Peroxynitrite can also oxidize protein, DNA, and lipids.
- .NO2 and peroxynitrite can interact with Tyr side chains in proteins to form nitrated proteins (Tyr-NO2), whose presence reflects inflammation.
Macrophages make use of RNIs in the killing of engulfed bacteria. How does one of humankinds greatest foes, Mycobacteria tuberculosis, the causative agent of tuberculosis, avoid this killing mechanism? It is estimated that the bacteria resides persistently in latent form in 2 billion people. If it becomes activated, it becomes one of the greatest killers. Consider the following table:
Killer Diseases through time (Scientist, June 2, 2003)
| Historic Pandemics | cases | deaths |
| Justinian Plaque (6th centr.) | 142 million (on 70% mort) | about 100 mill |
| China Plaque (Bubonic) 3rd Pandemic (1896-1930) | 30 milion | 12 milion |
| Spanish flu 1918-19 | 1 billion | 21 million |
Pandemics Today
| Pandemics Today | cases/yr | deaths/yr |
| Malaria | 300-500 mil | 1 million |
| TB | 8 mill | 2 mill |
| AIDS | 6 million | 3 mill |
People infected with the bacteria but without clinical symptoms must mount a sufficient enough immune response to restrain proliferation of the bacteria, but not enough to clear it from the body. Immune-compromised people (transplant recipients taking anti-rejection drugs or AIDS patients) have more difficulty in holding the bacteria at bay. One immune response mediator which restrains bacterial growth is the soluble protein interferon γ (a protein cytokine released by immune cells). This protein induces synthesis of nitric oxide synthase, producing .NO, which through the reactions listed above, can damage bacterial macromolecules.
Bacteria are "digested" in acidic phagosomes of the macrophage. Nitrite formed in the acid conditions from .NO (generated by inducible NO synthase) forms .NO2 and peroxynitrite which have antibacterial properties (better than anti-Tb drugs). Mutant mice that can not synthesize inducible NO synthase have little defense against the bacteria. Darwin e al. exposed the bacteria to sources of nitrite at pH conditions typical of macrophage phagosomes and found mutants sensitive to the RNIs. The mutations involved protein of the bacterial proteasome, a complex multi-protein complex which proteolyzes unwanted (presumably damaged) cellular proteins.
Bacterial genes encoding proteins associated with the bacterial proteasome seem to confer resistance to the effects of macrophage-inducted RNI production. The macrophage proteasome, like other eukaryotic proteasomes, is a cytoplasmic protein complex which degrades damaged cytoplasmic proteins. Although the mechanism is uncertain, the bacterial proteasome may rid the bacteria of nitrated or otherwise oxidized (damaged) proteins or remove the nitrate and facilitate refolding of the damaged protein.
![]() Peroxynitrite
in health and disease
Peroxynitrite
in health and disease
![]() Nitric
Oxide Synthases: Structure, Function, and Inhibition
Nitric
Oxide Synthases: Structure, Function, and Inhibition
Navigation
Return to Chapter 8A: The Chemistry of Dioxygen Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8A: The Chemistry of Dioxygen

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.