Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8 - OXIDATION/PHOSPHORYLATION
E: NITROGENASE - A REDUCTIVE USE OF METAL CENTERS
BIOCHEMISTRY - DR. JAKUBOWSKI
04/15/16
|
Learning Goals/Objectives for Chapter 8D:
|
E1. Nitrogenase: An Introduction
Beauty is in the eye of the beholder.
As the domain biochemistry covers the entire biological world, the extent of coverage of a given topic in textbooks can depend, in part, on the interest and experiences of the author(s) presenting the material. Is relevance a metric that should determine coverage? If so, books focused on human or medical biochemistry would surely omit photosynthesis. If topics are selected based on their importance for life, then photosynthesis must surely be covered. If so, then nitrogenase must also be included. If the degree of chemical difficulty for a chemical reaction and the amazing eloquence of the evolved biochemistry solution is considered, then both photosynthesis and nitrogen fixation must be presented. Even though nitrogen fixation is a reductive reaction, it shares strong similarities with the oxygen evolving complex of photosynthesis. They catalyze enormously important redox reactions that involve an abundant atmospheric gas using a very complicated and unique inorganic metallic cofactor that evolution has selected as uniquely suited for the job.
Every first year student of chemistry can draw the Lewis structure of dinitrogen, N2, which contains a triple bond and a lone pair on each nitrogen. If Lewis structures speak to them, they should be able to state that the triple bond makes N2 extraordinarily stable, thus explaining why we can breathe an atmosphere containing 80% N2 and not die. If they have taken biology, they are also aware that very few biological organisms can utilize N2 as a substrate, as this require breaking bonds between the nitrogen atoms, a chemical process reserved for nitrogen “fixing” bacteria found in rhizomes of certain plants. Lastly, they probably memorized that high pressure and temperature, in a process called the Haber-Bosch process, is used react N2 and H2 to form ammonia, NH3. As with any scientific advance, the Haber-Bosch process has brought both harm (its used for explosive weapons) and good (fertilizers). This process now fixes enough N2 in the form of fertilizers to support half of the world’s population, with nitrogenase supporting the rest. Effort is being devoted to genetically modify plants to make their own nitrogenase, eliminating the need for fertilizers but perhaps creating unforeseen problems of its own.
You might be surprised to find out that at room temperature the equilibrium constant favors ammonia formation, hence DG0 < 0. The reaction is favored enthalpically as it is exothermic at room temperature. It is disfavored entropically as should be evident from the balanced equation below: N2(g) + 3H2(g) → 2 NH3(g)
If the reaction is favored thermodynamically at room temperature, why doesn’t it proceed? This story sounds familiar as this same descriptor applies to oxidation of organic molecules with dioxygen. There we showed using MO theory that the reaction is kinetically slow. Same with NH3 formation. A superficial way to see this is that we must break bonds in the stable N2 to start the reaction, leading to a high activation energy, making the reaction kinetics sluggish.
One could jump start the reaction by raising the temperature, but that would slow an exothermic reaction. The Keq (or KD) and DG0 are obviously functions of temperature and for this reaction, and the reaction becomes disfavored at higher temperatures. The solution Haber found was high pressure, forcing the reaction to the side that has fewer molecules of gas, and high temperature to overcome the activation energy barrier and make the reaction kinetically feasible. A complex metal catalysts (magnetite - Fe3O4 -with metal oxides like CaO and Al2O3 which prevent reduction of the Fe with H2) provides an absorptive surface to bring reagents together and facilitate bond breaking in H2 and N2.
We have seen that nature has seemed to involved mechanisms involving the very strange oxygen evolving complex of Mn, Fe, S and Ca to oxidize another very stable and ubiquitous molecule, H2O. Now we explore the amazing mechanisms behind the nitrogenase complex which fixes N2 to form NH3.
What might be needed to drive this reaction biologically? You might surmise the list to include:
-
a source of energy, most likely ATP, to drive this difficult reaction and you’d be right;
-
a source of electrons as the N atoms move from an oxidation state of 0 in elemental N to 3- in NH3; this source turns out to be a protein called flavodoxin or ferridoxin. Of course, these electrons also have interesting sources before they were in the electron carriers of these proteins;
-
some pretty amazing metal centers to accept and donate electrons in a controlled way; these centers are mostly FeS clusters with an additional cluster containing molybdenum (Mo). The clusters are named F, P, and M
-
a source of hydrogen; you might have guessed correctly that it’s not H2 gas (from where would that come?), but H+ ions which are pretty ubiquitously available.
-
a net reaction that is different that the Haber-Bausch process (N2 + 3H2 → 2 NH3).
Here is the actual reaction catalyzed by nitrogenase:
N2 + 8e- + 16ATP + 8H+ → 2NH3 + H2 + 16ADP + 16Pi.
Let’s think a bit about the reaction. As electrons are added the attractions between the nitrogen atoms must decrease. Eventually bonds between them must be broken. Protons could be easily added to maintain charge neutrality. A basic mechanism might involve intermediates as shown below:
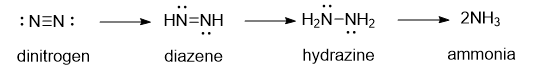
Nitrogenase can also other small molecules with triple bonds, including :C=O: and H-C=C-H.
E2. The Structure of Nitrogenase
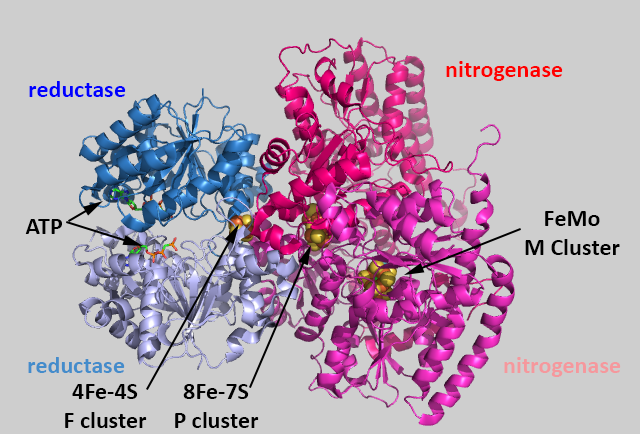
Nitrogenase is a multiprotein complex containing the following:
-
Two identical subunits, E and F, which have binding sites for the mobile carrier of electrons (the protein ferrodoxin or flavodoxin), ATP, and an FeS cofactor (4Fe-4S, called the F cluster) which accept electrons. These subunits are hence called the nitrogenase reducatase subunits
-
a heterodimer of an alpha and beta subunits. The a (alpha chain) binds the 8Fe-7S F cluster and the Fe-S-Mo M cluster. These subunits comprise the (di)nitrogenase subunits
The overall structure of the protein complex with bound ATP and metal centers are shown below.
An interactive Jsmol structure of nitrogenase is found in the link below.
![]() JSMol (HTML5)
JSMol (HTML5)
Protopedia: Nitrogenase
An enhanced view of the bound cofactors and ATP are shown in the same spatial orientation in the figure below.
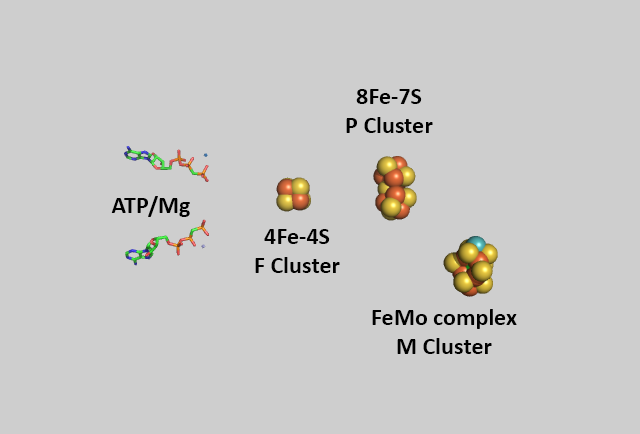
The metal centers are shown below in more detail in both line and space fill views.
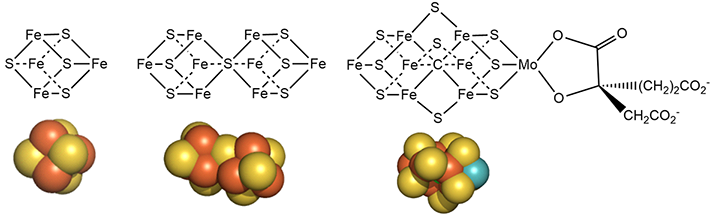
Mo is bound to 3 sulfur ions and two a OH and carboxyl group of 3-hydroxy-3-carboxy, adipic acid in the crystal structure of shown above.
The M cluster has an interstitial carbide ion that derives from -CH3 attached to the sulfur of S-adenosyl-methionine (SAM) allowing the carbide to be labeled with either 13C or 14C for mechanistic studies. These labeled carbides are not exchanged or used as a substrate when the enzyme undergoes catalytic turnover. Hence it seems that the carbide probably just stabilizes the M cluster. it won't be shown in the figures below showing more detailed mechanisms.
E3. Nitrogenase Reaction: Part 1 - Addition of Electrons and Protons
The sequential path of electrons from the reductase subunit containing the F cluster to the P and M clusters in the nitrogenase subunit should be apparent from the figures above. We will concentrate on the binding of N2 and how it receives electrons from the M cluster. The figure below shows the FeMo-cofactor and some adjacent amino acid residues. The Mo is not shown in space fill.
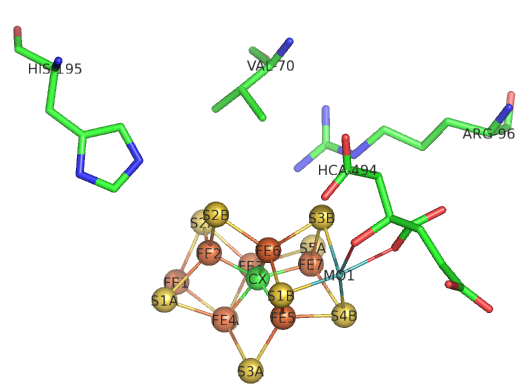
Question 1: If Val 70 is mutated to Ile, substrate appears not to access the cluster. Which part of the cluster most likely interacts with N2?
Question 2: His side chains are often found at enzyme active sites. What role might they play in catalysis?
The Lowe and Thorneley (LT) model has been proposed as a mechanism for dinitrogen reduction. In this model an electron and proton are added to the oxidized form of the enzyme (Eo) to produce E1. This is repeated 3 more times to form sequentially, E2, E3 and E4. Only then does N2 bind and the reduction of N2 occur. Two of the added electrons are accepted by H+ ions which form H2, which is liberated on N2 binding.
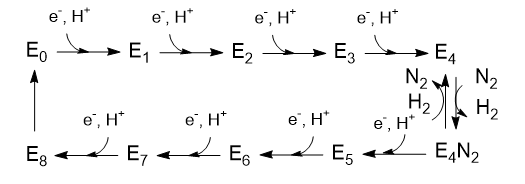
Question 3: Based on oxidation numbers of N and H in N2, NH3, H+ and H2 and the mechanism above, would you expect 8 electrons to be needed?
The crystal structure shows 2 ATPs bound to the reductase subunit. The stoichiometry of the reaction shows 16 ATP used. Simple math suggests that 2 ATP are cleaved to support the entry of one electron into the complex.
Part 1 - E1-E4: A potential structure for the E4 intermediate is shown below. Note the carbide is not shown. This is often called the Janus intermediate as it is half-way through the catalytic cycle. It is named for Janus, the Roman god of beginnings and transitions, and has been ascribed to gates, doors, doorways and passages. Janus is typically shown with two faces, one looking to the future and one to the past.
Question 4: How can you tell that this figure represents E4?
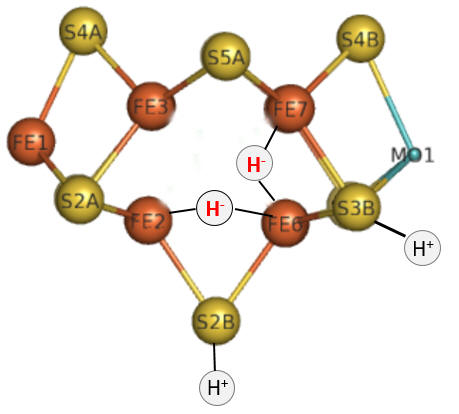
The hydrides bridge 2 Fe ions so these are examples of three-center, two-electron bonds.
How does this reaction occur? We must look to organometallic chemistry to help us understand the mechanism of this and subsequent steps. Clearly a hydride equivalent has been added to the metals, associated with the oxidation of metal ions in the center. This particular reaction is called an oxidative addition. Presumably the sulfur ions act as Lewis bases as they gain protons from a Lewis acid, probably HIs 195.
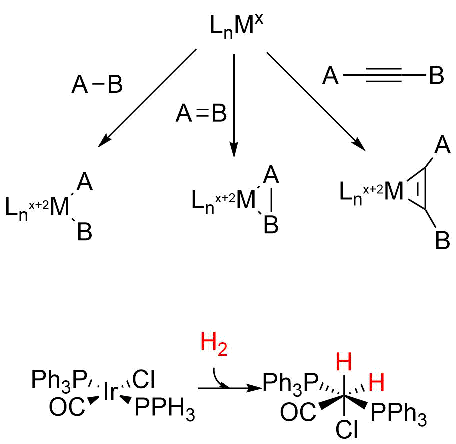
Oxidative addition reactions
A figure showing oxidative additions for three different types of reactants are shown below. Note the specific example for insertion of H2, an example somewhat similar to the hydride additions to the M cluster. Oxidative reduction occurs most readily when the two oxidation states of the metal ion are stable. It is likewise favored for metal centers that are not sterically hindered (makes sense if A-B is to be added) and if A-B has a low bond dissociation energy.
Question 5: What role might the H+ ions have in the E4 complex shown above? Why might they be relatively close to the hydrides?
One way to study reaction intermediates is to trap them. If N2 can't access the binding site and the temperature is reduced, the accumulated hydrides and H+ in E4 might interact as the reaction goes back to E1.
Question 6: What might be a likely product in such a case? What mutant might be used to attempt this experiment.
E4. Nitrogenase Reaction: Part 2 - Reduction of N2
Step E4 to E5 seems a bit bizarre as H2 gas is released. This would seem to waste ATP but it must be obvious that we should trust evolution which has created this enzyme. Do you think that this reaction would be facilitated by having the hydrides on one Fe ion, Fe6? The mechanism is another classic organometallic reaction, reductive elimination, the reverse of oxidative addition.
Reductive elimination reactions
In this reaction, a molecule is eliminated or expelled from the complex as the metal ion is reduced and adds two electrons. The figure below shows reductive elimination. Reductive elimination occurs most readily in higher oxidation state metal centers which can be stabilized on reduction. It occurs most readily from electron rich ligands and if the other surrounding ligands are bulky. The dissociating species must also be cis to each other as they form a bond to each other when they leave.
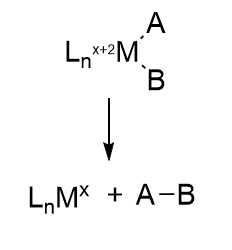
Oxidative addition and reductive elimination at metal centers are often coupled together in organometallic catalytic cycles with some rearrangement or other modification occurring between them. Think about it. The FeS clusters must return to their original oxidation state after the complete LT cycle. We will encounter another organometallic reaction after the addition of N2, migratory insertion, in the second half of the reaction.
Is H2(g) really released? To study this, investigators have used an alternative substrate, acetylene, HC=CH, in the presence of D2 and N2 in an aqueous system.

The acetylene was reduced and formed C2H2D2 and C2H3D. Hence E4 must have had 2 Ds in it, and E2 probably 1. These results support the reversible reductive elimination mechanism for the E4 to E4:N2 reaction above. Previously it had been shown the H+ are reduced by D2 in the presence of D2 and N2 in an aqueous system, so these results are consistent. In additional support, deuterium from D2 is not incorporated into products (C2H2D2, C2H3D or HD) in the absence of N2.
Let's return for a moment to the bridging hydrides as shown again in the figure below.

To form H2, the H- hydrides must be protonated.
Question 5: The hydrides in the model above are shown as bridging (involved in 3 center, 2 electron bonds) and not as terminal (as shown for the H+). Which would be more resistant to protonation and hence reductive elimination, bridging or terminal hydrides?
The bridging hydrides are now stable enough to react with the desired substrate, N2. The M cofactor is large enough to create a face of four Fe ions (see figure below) which are coordinated to a carbide on the bottom face. To create this face of Fe ions requires a cluster of sufficient size with the 6 Fes and 1 central C forming a trigonal prismatic geometry.

E5. Oxidation States of Nitrogenase Fe centers
First Half: It would be difficult to assign specific oxidation states to each Fe ion in the M complex. Instead we can assign relative changes in the oxidation states as the reaction proceeds from E0 to E4. In each step of the LT model, 1 electron is added. We will first assign this to a average Fe ion, M0, with an arbitrarily assigned oxidation state of 0. On addition of 1 electron, the oxidation state would go from M0 to M-1 as the metal is reduced. The M-1 state is then oxidized as an electron is transferred to H+, and when 2 electrons are transferred, and a single H- is made. The diagram below shows the change in oxidation state in going from E0 to E4.
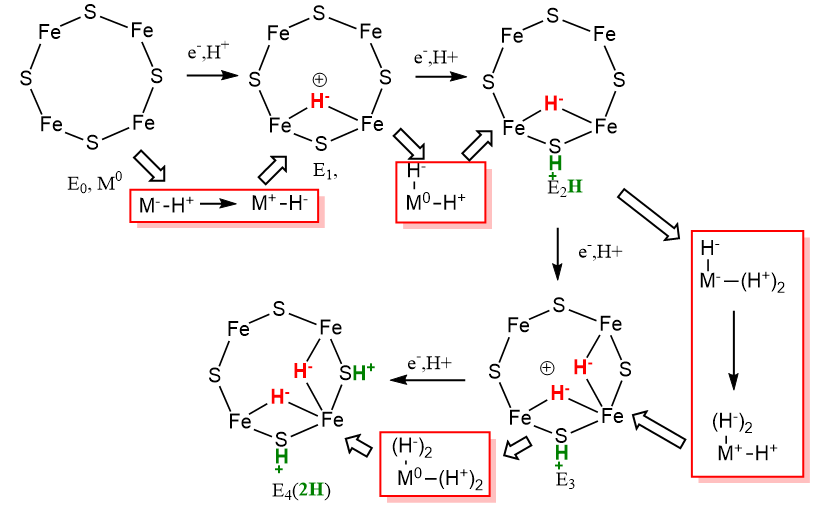
The red boxes highlight thermodynamic cycle-like steps which show how the changes in redox state of the Fe ions (M) could be visualized. Note that in going from E0 to E4, the actual oxidation state of M changes from 0 to +1 to 0 to +1 and back to 0. That is quite amazing given that 4 electrons have been added. Note also that in the red box going from step E0 to E1, M goes from -1 to +1 which corresponds to our description of an oxidative addition when the metal center looses two electrons. This mechanism shows that nitrogenase could be considered a "hydride storage device".
Second Half (facing forward to production NH3):
How does N2 initially interact with E4? It must depend on how the hydrides are released as H2, which evidence shows occurs by reductive elimination (re) and not hydride protonation (hp). On addition, the N2 very quickly is converted to diazene, HN=NH, with the departing H2 taking with it 2 H+s and 2 electrons (or reducing equivalents). These events could occur as shown in the figure below.
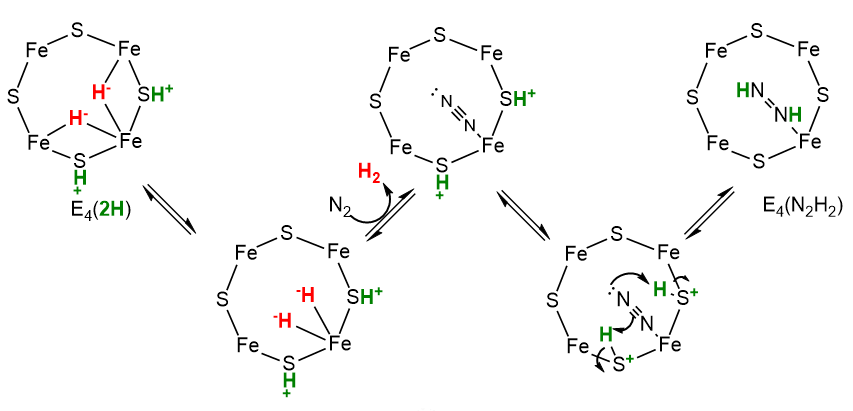
Now, with N2 bound as diazene (N2H2) and H2 released, the rest of the reaction could occur as shown below. One new step, a migratory insertion, is shown.
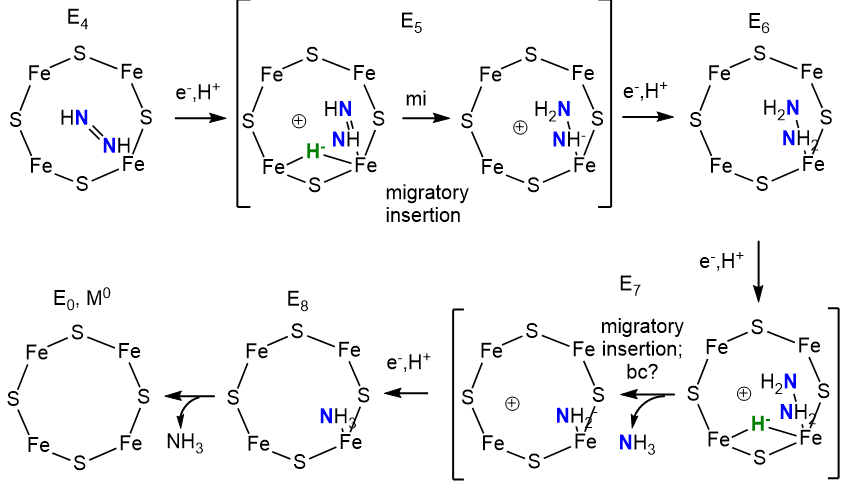
The two halves of the reaction are similar with bridging hydrides utilized. The E4 Janus intermediate links the two halves together.
E6. Links and References
Brian M. Hoffman et al. Mechanism of Nitrogen Fixation by Nitrogenase: The Next Stage, Chem. Rev. 2014, 114, 4041−4062 dx.doi.org/10.1021/cr400641x
Jared A. Wiig et al. Tracing the Interstitial Carbide of the Nitrogenase Cofactor during Substrate Turnover. J Am Chem Soc. 2013 April 3; 135(13): 4982–4983. doi:10.1021/ja401698d
Return to Chapter 8D: The Light Reactions of Photosynthesis Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8D: The LIght Reaction of Photosynthesis

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.