Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8 - OXIDATION/PHOSPHORYLATION
D: THE LIGHT REACTION OF PHOTOSYNTHESIS
BIOCHEMISTRY - DR. JAKUBOWSKI
04/15/16
|
Learning Goals/Objectives for Chapter 8D: After class and this reading, students will be able to
|
D1. Introduction
"Of all the biochemical inventions in the history of life, the machinery to oxidize water — photosystem II — using sunlight is surely one of the grandest." (Sessions, A. et al, 2009)
We have just seen how we can transduce the chemical potential energy stored in carbohydrates into chemical potential energy of ATP - namely through coupling the energy released during the thermodynamically favored oxidation of carbon molecules through intermediaries (high energy mixed anhydride in glycolysis or a proton gradient in aerobic metabolism) to the thermodynamically uphill synthesis of ATP. There is a situation that occurs when we wish to actually reverse the entire process and take CO2 + H2O to carbohydrate + O2. This process is of course photosynthesis which occurs in plants and certain photosynthetic bacteria and algae. Given that this process must by nature be an uphill thermodynamic battle, let us consider the major requirements that must be in place for this process to occur:
- A strong oxidizing agent must be formed which can take water and oxidize it to dioxygen. We know that redox reactions occur in the direction of stronger to weaker oxidizing agent (just as acid base reactions are thermodynamically favored in the direction of strong to weak acid). Somehow we must generate a stronger oxidizing agent than dioxygen, which often has the most positive standard reduction potential in tables.
- Plants must have high concentrations of a reducing agent for the reductive biosynthesis of glucose from CO2. The reducing agent used for most biosynthetic reactions in nature is NADPH, which differs from NADH only by the addition of a phosphate to the ribose ring. This phosphate differentiates the pool of nucleotides in the cells used for reductive biosynthesis (NADPH/NADP+) from those used for oxidative catabolism (NADH/NAD+)
- Finally, plants need an abundant source of ATP which will be required for reductive biosynthesis.
We will discuss only the light reaction of photosynthesis which produces these three types of molecules. The dark reaction , which as the name implies can occur in the dark, involves that actual fixation of carbon dioxide into carbohydrate using the ATP and NADPH produced in the light reaction.
D2. Photoexcitation and Electron Transfer
Obviously, the energy to power the light reactions comes directly from sunlight. Clue two is that plants have an organelle that animal cells don't - the chloroplast. Its structure is in many ways similar to a mitochondria in that it has internal membranes (thylacoid membranes) surrounding enclosed compartments.
Plants have many pigments (chlorophyll, phycoerthryins, carotenoids, etc.) whose absorption spectra overlap that of the solar spectra. The main pigment, chlorophyll, has a protophorphryin IX ring (same as in heme groups) with Mg instead of Fe. When the chlorophyll absorbs light, the excited electrons must relax eventually to their ground state. It can do this by either radiative or nonradiative decay. In radiative decay, a photon of lower energy is emitted (after some energy has already been lost by vibrational transitions) in a process of either fluorescence or phosphorescence. In nonradiative decay, the energy of an excited electron can be transferred to another similar molecule (in this case other chlorophyll molecules) in a process which excites the energy of an electron in the second molecule to the same excited state. (It is as if a photon is released by the first excited molecule which then is absorbed by an electron in a second molecule to excite it to the same exited state, although the excitation occurs without photon production). In this fashion, energy is transferred from one chlorophyll to another. This type of energy transfer is called resonance energy transfer or exciton transfer.
Figure: resonance energy transfer
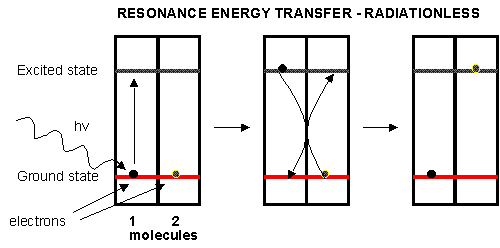
One type of chlorophyll has slightly different characteristics, however. Because of its unique environment, the energy level of the excited state electron is lower than in the rest of the chlorophyll molecules, in much the same way that pKa's of amino acid side chains differ with environment, and the standard reduction potential of FADs that are tightly bound to enzymes differ due to the different environment of FAD/FADH2 These unique chlorophyll centers are called reaction centers.
Figure: reaction centers
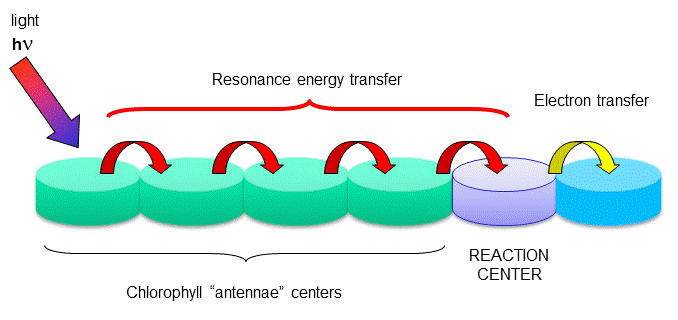
The rest of the chlorophyll molecules act as antennas which transfer energy to the reaction centers.
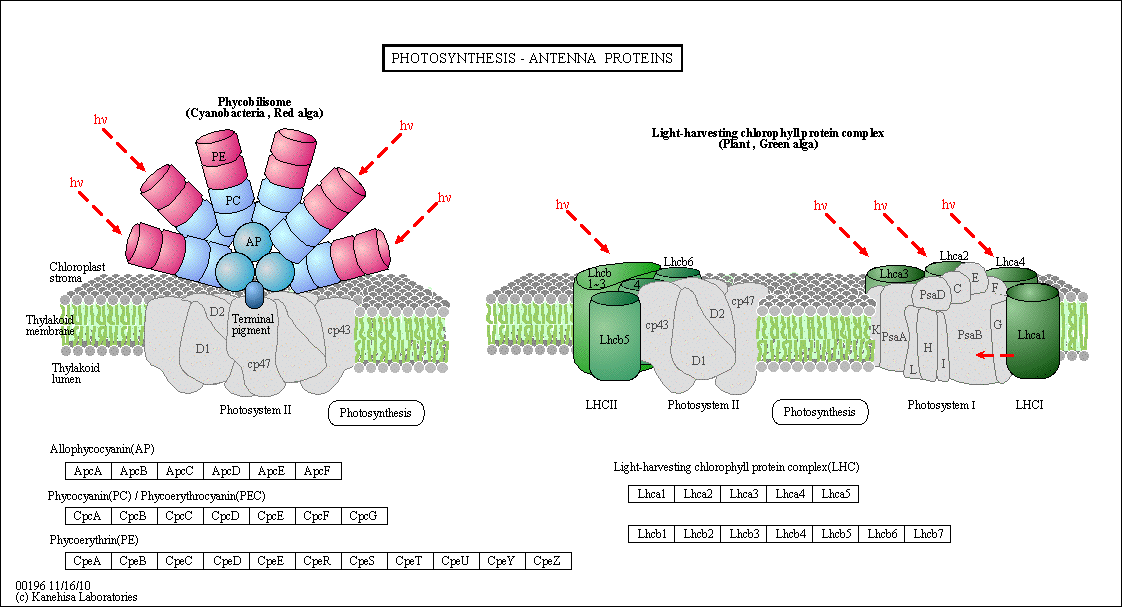
Figure: Antennae Proteins
(reprinted with permission from Kanehisa Laboratories and the KEGG project: www.kegg.org )
An electron in an adjacent excited state chlorophyll (which is at a higher level than the excited state energy of the reaction center) can then be transferred to this lower energy state level in the reaction center, in a process which forms a positively charged ion from the first excited state molecule and an anion from the recipient reaction center. This process of energy transfer is called electron transfer.
Figure: electron transfer
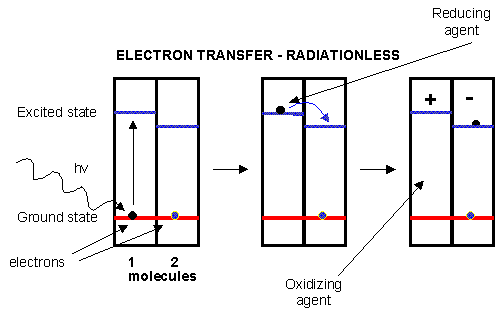
D3. The Light Reactions of Photosynthesis
Photoexcitation of the non-reaction center chlorophyll turns that molecule into a good reducing agent, which transfers its electron to the nearest excited state level of the reaction center chlorophyll. If you count both step together, the non-reaction center chlorophyll gets "photooxidized", in the process producing the "strong" oxidizing agent which is the positively charged chlorophyll derivative. The extra electron passed onto the second molecule will eventually be passed on to NADP+ to produce NADPH. The light reaction of photosynthesis in green plants is shown below. In this process, in a scheme that is reminiscent of electron transport in mitochondria, water is oxidized by photosystem II. Electrons from water are moved through PSII to a mobile, hydrophobic molecule, plastaquinone (PQ) to form its reduced form, PQH2. PSII is a complicated structure with many polypeptide chains, lots of chlorophylls, and Mn, Ca, and Fe ions. A Mn cluster, called the oxygen evolving complex, OEC, is directly involved in the oxidation of wate. Two key homologous 32 KD protein subunits, D1 and D2, in PSII are transmembrane proteins and are at the heart of the PSII complex. Another photosystem, PS1, is also found further "downstream" in the electron transport pathway. It takes electrons from another reduced mobile carrier of electrons, plastocyanin (PCred) to ferredoxin, which becomes a strong reducing agent. Ferrodoxin is a protein with an Fe-S cluster (Fe-S-Fe-S in a 4 membered ring, with 2 additions Cys residues coordinating each Fe). It ultimately passes its electrons along to NADP+ to form NADPH. A summary of the light reaction in plants and standard reduction potentials of the participants, are shown below. Note that many of the complexes produce a transmembrane proton gradient. In contrast to mitochondria, the lumen (as compared to the mitochondrial matrix) becomes more acidic that the other stroma. Protons then can move down a concentration gradient through the C0C1ATPase to produce ATP required for reductive biosythesis of glucose.
Figure: THE LIGHT REACTIONS OF PHOTOSYNTHESIS
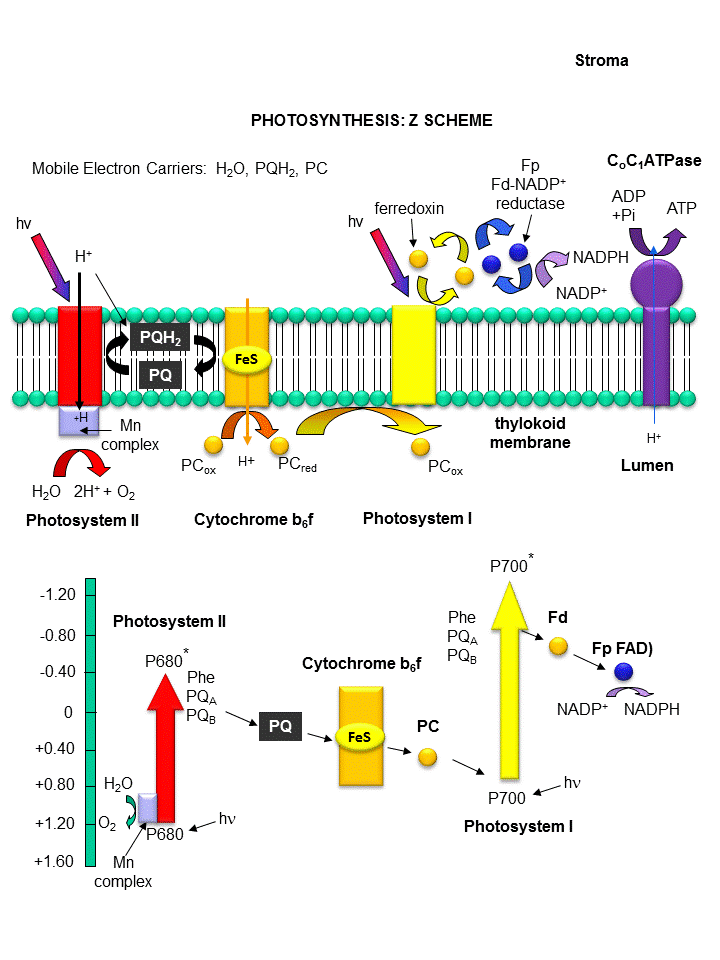
Figure: Detailed View of Light Reaction of Photosynthesis (reprinted with permission from Kanehisa Laboratories and the KEGG project: www.kegg.org )
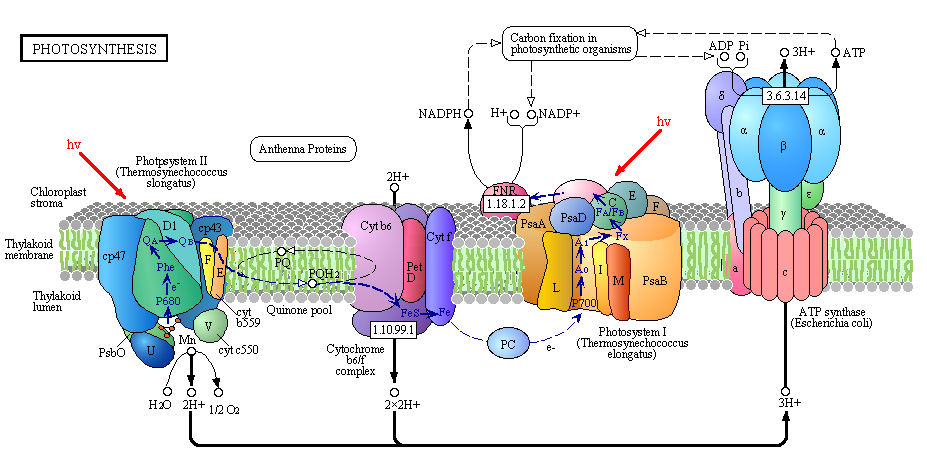
Boxed number represent Enzyme Commission Number. Original KEGG Map with imbedded links.
D4. Photosystem II
The net reaction carried out by PS2 is the oxidation of water and reduction of plastoquinone.
PQ + H2O --> PQH2 + O2 (g)
Note that water is not converted to 2H2 + O2 , as in the electrolysis of water. Rather the Hs are removed from water as protons in the lumen of the cholorplast, since the part of PSII which oxides water is near the lumenal end of the transmembrane complex. Protons required to protonated the reduced (anionic) form of plastaquinone to form PQH2, an activity of PSII found closer to the stroma, derive from the stroma. which then can be used to protonated the "anionic" form of reduced PQ to form PQH2.
A quick look at standard reduction potentials shows that the passing of electrons from water (dioxygen SRP = +0.816 V) to plastoquinone (approx SRP of 0.11 ) is not thermodynamically favored. The process is driven thermodynamically by the energy of the absorbed photons.
An early crystal structure of PSII from a photosynthetic cyanobacterium showed that it consists of 17 polypeptide subunits with metal and pigment cofactors and over 45,000 atoms. (Zouni, Nature, 409, 739, 2001). Of particular interest is the P680 chlorophyll reaction center, which consists of four monomeric chlorphylls adjacent to a cationic Tyr-D side chain which destabilizes the chlorophyll molecules. When H2O gets oxidized to form dioxygen, 4 electrons must be remove by photoactivated P680. In PSII, this process occurs in 4, single electron steps, with the electrons first being transferred to a Mn4 cluster cofactor (of composition Mn4Ca1Cl1-2(HCO3)y. This inorganic Mn cluster is often called OEC (oxygen evolving complex) or WOC (water oxidizing complex). The electrons passed through the Mn complex are delivered to P680 by a photoactive Tyr free radical (Tyr Z). The actual structure of the OEC could not be resolved in this structure, but other structural and spectroscopic data support the structure below (Chem. Rev., 2001, 101, 21-35), which also shows a possible mechanism for electron and proton transfer from water to form dioxygen. 5 discrete intermediates of the OEC, S0-S4, are suggested from the experimental data (Kok cycle). These were postulated from experiments in which spinach chloroplast were illuminated with short light pulses. A pattern of dioxygen release was noted that repeated after 4 flashes. Ultimately, light absorption by P680 forms excited state P680* which donates an electron to pheophytin (which passes them to quinones) to form P680+, which receives electrons from the OEC, specifically the TyrZ radical. As we will see in a bit, this structure and mechanism has been called into question by new crystallographic structures.
Figure: OEC - Mechanism of Water Oxidation
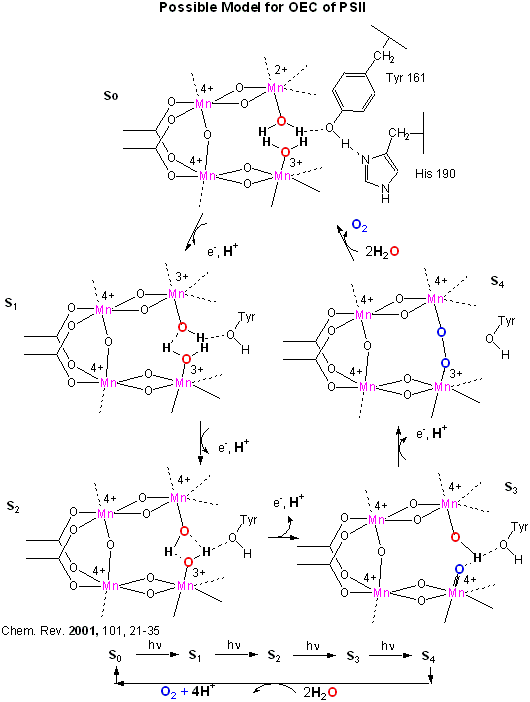
Investigators have made non-peptide mimetics of superoxide dismutase to facilitate therapeutic removal of excess superoxide formed in brain and heart tissue. These may arise after an oxidative burst from reperfusion of these tissues after heart or brain attacks. Likewise, scientist are trying to build synthetic PSII-OEC complexes which could be used to form dioxygen or hydrogen gas for fuels.
In summary, for PSII in plants
- a pair of chlorophylls (P680) in the D subunits absorb light (maximum absorbance around 680 nm) and reach an excited state
- electron transfer from P680 to a nearby chlorophyll with a lower energy level for the excited state electron occurs. This chlorophyll has 2 H+ ions in the chlorophyll instead of Mg2+occurs. The P680 now becomes cationic, P680+.
- This "anionic" chlorophyll transfers an electron to oxidized plastoquinone.
- The P680+, a strong oxidizing agent, removes one electron from H2O-OEC complex.
Steps 1-4 repeat three more times, each requiring another photon and each cycle producing another electron which passes on through the system. Remember that when O2 acts as an oxiding agent, it gains four electrons. The first produces superoxide, the next peroxide, and two more produce oxide which when protonated is water. Hence two waters and four cycles are required to remove the four electrons required to produce dioxygen.
A similar mechanism is found in PSI, except plastacyanin, not dioxygen is oxidized, with electrons moved to ferrodoxin. This is likewise a difficult process since the reduction potential for oxidized plastocyanin (the form that can act as a reducing agent) is +0.37 while for ferrodoxin it is -0.75. This transfer of electrons is an uphill thermodynamic battle since the more positive the standard reduction potential, the better the oxidizing agent and the more likely the agent becomes reduced. What drives this uphill flow of electrons. Of course, it is the energy input from the photon.
![]() Jmol:
Updated Light Harvesting
Complex from Spinach 1RWT
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Light Harvesting
Complex from Spinach 1RWT
Jmol14 (Java) |
JSMol (HTML5)
![]() Jmol: Updated
Detailed Photosystem II from S. elongatus
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
Detailed Photosystem II from S. elongatus
Jmol14 (Java) |
JSMol (HTML5)
![]() Jmol: Updated
Photosystem I: A Photosynthetic Reaction Center and Core Antenna System From
Cyanobacteria 1JBO
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Updated
Photosystem I: A Photosynthetic Reaction Center and Core Antenna System From
Cyanobacteria 1JBO
Jmol14 (Java) |
JSMol (HTML5)
D5. The Oxygen Evolving Complex (OEC)
The crystal structure of PS2 from T. vulcanus has significantly improved our understanding of the OEC and electron flow on water oxidation.Because of its similarity to other pathways you have studied, we will concentrate on developing an understanding of the amazing Photosystem II from Thermosynechococcus vulcanus, a cyanobacteria (19 subunits with 35 chlorophylls, two phenophtyins, 11 beta cartotenes, 2 plastoquinones, 2 heme irons, 1 non heme iron, 4 Mn ions, 3-4 Ca ions, 3 Cl ions, 1 carbonate ion, and around 2800 water molecules).
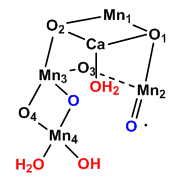
Nature has appeared to evolve a single gene for the part of PSII that binds an inorganic cluster involved in oxygen formation. The cluster is called the Oxygen Evolving Complex (OEC) and is bound to a central protein in PS2. The cluster, Mn4CaO5, appears identical in all photosynthetic organisms, and is shown below. Researchers were surprise to find that the Ca ion was an integral part of the basic geometric “framework” of the OEC instead of a Mn which was found to be “dangling” from the basic geometric framework.
Figure: OEC structure from T. vulcanus
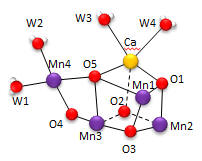
The shape outlined by O5-Ca-O1-Mn1 and Mn3-O2-Mn2-O3 is similar to what organic molecule? Describe how every two adjacent Mn are linked. Describe the coordination of Ca. Four waters (W1-W4) are coordinated to the OEC. No others were found in the region. What role may some of the waters have in the complex?
The amino acid side chains that interact with the OEC are shown below. The coordination number for all the Mn ions (including interacting waters) is identical. What is it for Mn1? For Ca? (Note that the Ala 344 is at the carboxy terminus of one of the protein subunits.)
Figure: OEC and surrounding amino acids from T. vulcanus
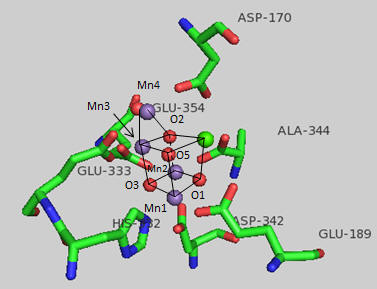
The OEC can be thought of as a distorted chair with a Mn3-O4-Mn4-O5 back. Bonds to O5 are longer than the other bonds. What might that imply about the strength of the bonds compared to the other metal-oxygen bonds? What is the expected charge of the oxo ligands in the OEC? If the metal-O5 bond lengths are longer than the others, what might that imply about the charge on O5? Would it be consistent with a ligand other than an oxo ligand? What might it be?
Which waters are most proximal to O5? Ultimately an O-O bond must form between two waters. Which are the most likely candidates based on the structure above? Which ion, Ca or Mn, would most likely be involved in electron transfer to water oxygens? Write the electron configuration for both ion. Standard reduction potentials for Mn are shown below. What is the oxidation number of Mn in each compound. Which oxidation state might be sufficient for the oxidation of H2O in the OEC?
- Mn2+ (aq) + 2 e-→ Mn (s) -1.185
- MnO4- (aq) + 2 H2O (l) + 3 e- → MnO2 (s) + 4 OH- 0.595
- MnO2(s) + 4H+ + e- → Mn3+ + 2H2O +0.95
- MnO2(s) + 4H+ + 2e- → Mn2+ + 2H2O +1.23
- MnO4- (aq) + 8 H+ (aq) + 5 e- → Mn2+ (aq) + 4 H2O (l) 1.507
- MnO4- (aq) + 4 H+ (aq) + 3 e- → MnO2 (s) + 2 H2O (l) 1.679
- HMnO4- + 3H+ + 2e- → MnO2(s) + 2H2O +2.09
- O2(g) + 4H+ + 4e- → 2H2O +1.229
The first structure of OEC above shows the two di-m-oxybridges that connect Mn4 to Mn1 and Mn3. This might give us clues as to the mechanism of water oxidation by the complex. To form O2, two water molecules are required as substrates. Presumably they bind as waters between adjacent Mns . What reaction intermediates may be bound to the Mn as water converts to O2? How many electrons are required for the complete oxidation?
D6. Water Oxidation - The Kok Cycle
The mechanism by which water is oxidized occurs through a 5 stage Kok cycle illustrated below?
Figure: Kok Cycle
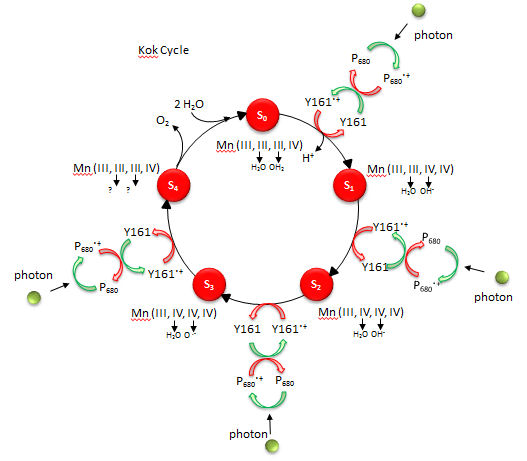
The Sn states in the Kok diagram denote different discrete oxidation states where n the number of oxidative “equivalent” stored in the OEC during cycle progression.
What happens to Mn ions in the OEC during the cycle? What is required to initiate the next stage of the cycle starting from So? What is the role of Y161.+, an amino acid in PS2 that is very close to the OEC as shown in the figure below?
Figure: Y 161 and the OEC
A new mechanism has been proposed based on the recent crystal structure. A possible model for S3 is shown below with the distorted cubane structure and with an oxy and oxyl ligands. Add a curved arrow to show how an oxygen-oxygen bond is made.
Figure: Model for S3 of Kok Cycle
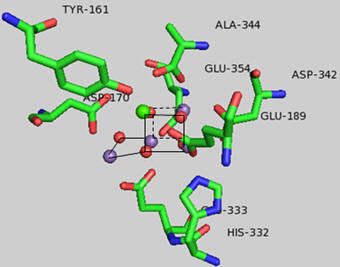
Given the maximal packing of ligands (including water) around the OEC, there must be a way for water to enter the site and for protons that are removed to move away from the complex and toward the lumen where they would contribute to the development of a proton gradient.
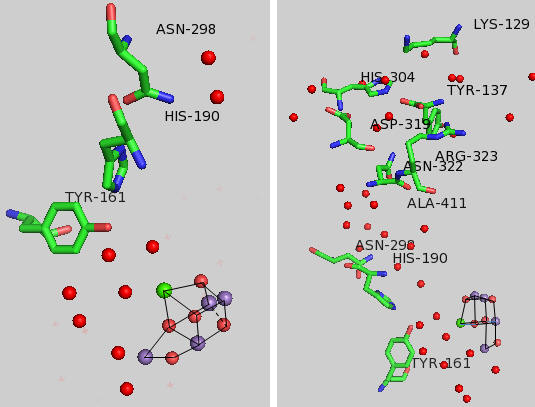
Tyr 161 is a critical amino acids that is situated proximal both to the OEC and to the source of electrons that reduce water. The figure below shows the immediate environment around Y161 and the OEC. Describe the role of the groups shown in the figure below. An expanded view of the OEC and part of the protein is shown to the right. Tyr 137 lies near the luminal interface of the protein complex. Describe the figure below and the role of the groups shown.
Figure: Environment of Y161 in the OEC of T. vulcanus
D7. Chlorophylls and the Reaction Center
Photosynthetic bacteria and plant have an abundance of molecules that interact with visible light. The main one found in PS2 is chlorophyll a whose structure is shown below.
Figure: Chlorophyll a
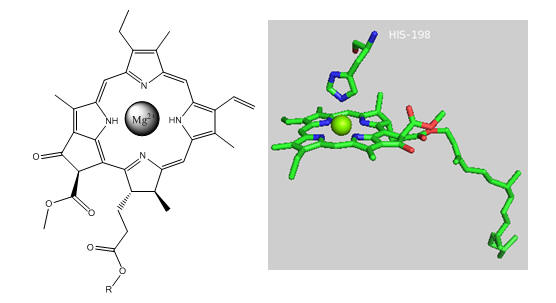
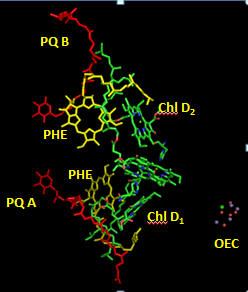
In PS2 there are 35 chlorophylls. Seven of them use water for ligand to the Mg ion, while the rest use His (as shown in the example above). Why might there be so many chlorophylls in PS2? The chlorophylls (Chl) are arranged in a very specific arrangement as shown below.
Figure: PS2 reaction center and antennae chlorphylls - overview
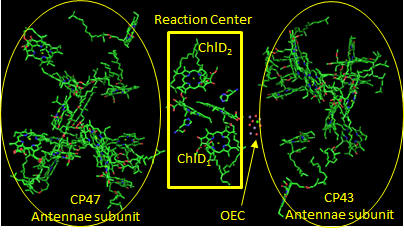
Two special chlorophylls, ChlD1 and ChlD2, accompanied by partner chlorophylls, PD1 and PD2 that are coplanar to each, are found near the OEC and are the key chlorophylls that turn OEC into a powerful enough oxidant to oxidize H2O. These 4 chlorophylls are called the reaction center. What might be the role of chlorophylls in the “antennae” subunit “activating” the reaction center?
Now we will consider the key process: how the reaction center chlorophyll becomes nature’s most powerful oxidant.
Figure: Reaction Center T. vulcanus
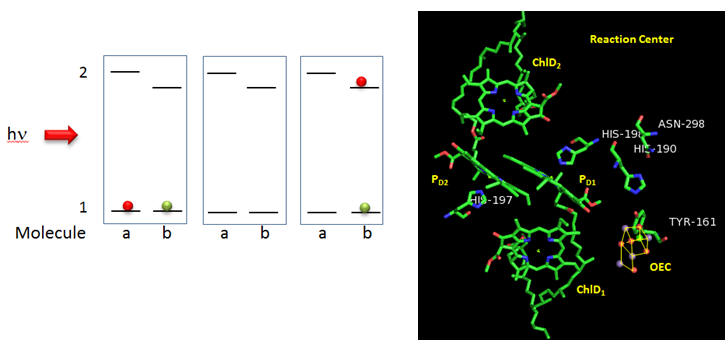
Why might molecule “b” have a lower excited state energy? Show by adding arrows and moving the given electrons in the above diagram how excitation of molecule a can lead to the final state shown to the right. What would be the charge on molecule “a”? Molecule “b”? Which would might be a powerful oxidant?
Next we must consider how the oxidant made at the reaction center, denoted as P680.+ leads to changes in the OEC as illustrated in the S0-4 oxidative intermediates in the Kok cycle.
Figure: Y161 Radical Formation
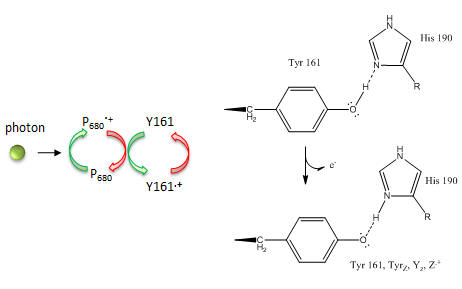
Based on the figures above, which chlorophyll in the reaction center, ChlD1 or ChlD2 represents molecule “a” in the top left figure? Molecule “b”? What might be the role of the coplanar partner molecules PD1 and PD2 ?
In reality, quantum mechanical calculations show a coupling between ChlD1 or ChlD2 such that 80% of the positive charge and radical character is situated on ChlD1 and 20% is on ChlD2.) How does Y161.+ lead to sequential transfer of “oxidizing” equivalents to the OEC? How is Y161 regenerated so that catalysis is continued?
The last feature to consider is the fate of second member of the reaction pair represented by molecule “b” to the right. It must also be regenerated. What must is lose to be regenerated?
Cofactors bound in the reaction center allow a path for electron flow. These include pheophytins (PHE) and plastoquinones A and B. Pheophytins are chlorophyll a molecules without the central Mg ion. The structure of plastoquinone is show above. Trace a likely path for electron flow from molecule “b” to Phe, PQA and then to PQB (with the realization that quantum mechanical coupling and tunneling occur in this process so that the process are coupled).
Figure: Cofactors in Reaction Center PS2 T. vulcanus
D8. Plant Protection
Plants have evolved a great ability to absorb light over the entire visible range of the spectra. Can they absorb to much energy? The answer is yes, so plants have developed many ways to protect themselves. IF too much light is absorbed, the pH gradient developed across the thylacoid membranes becomes greater. This is sensed by a protein, PsbS, and through subsequent conformational changes transmitted through the light-harvesting antennae, the excess light energy is dissipated as thermal energy. Mutants lacking PsbS showed decreased seed yield, a sign that it became less adaptable under conditions of stress (such as exposure to rapidly fluctuating light levels). Molecules called xanthophylls (synthesized from carotenes - vit A precursors) such as zeaxanthin are also important in excess energy dissipation. These molecules appear to cause excited state chlorophyll (a singlet like excited state dioxygen) to become deexcited. Without the xanthophylls, the excited state chlorophyll could deexcite by transfer of energy to ground state triplet dioxygen, promoting it to the singlet, reactive state, which through electron acquisition, could also be converted to superoxide. These reactive oxygen species (ROS) can lead to oxidative damage to proteins, lipids and nucleic acids, alteration in gene transcription, and even programmed cell death. Carotenoids can also acts as ROS scavengers. Hence both heat dissipation and inhibition of formation of ROS (by such molecules as vitamin E) are both mechanism of defense of excessive solar energy
Given that both plants and animals must be protected from ROS, antioxidant molecules made by plants may prove to protect humans from diseases such as cancer, cardiovascular disease, and general inflammatory diseases, all of which have been shown to involve oxidative damage to biological molecules. Humans, who can't synthesize the variety and amounts of antioxidants that are found in plants, appear to be healther when they consume large amounts of plant products. These phytomolecule also have other properties, including regulation of gene transcription which can also have a significant effect on disease propensity.
D9. Production of Hydrogen: Hydrogenases (repeated from 8B)
Our world desperately needs an energy efficient way to produce H2 for energy production without producing waste pollutants. Catalytic cracking of molecules and newly developed fuel cells offer two possibilities. Wouldn't it be great if a reactant like water could be used for H2 production (without the use of electrolysis) or expensive metal catalysts? Nature may show the way. Bacteria (even E. Coli found in our GI system) can use simple metals like iron to produce H2 from H+ with electrons for the reduction of H+ coming from a donor (such as a reduced heme in proteins):
Dred+ H+ <=> Dox + H2
The reaction is also reversible in the presence of an acceptor of electrons from H2 as it gets oxidized:
Aox+ H2 <=> Ared + H+
The enzymes that catalyze hydrogen production are hydrogenases (not dehydrogenases). Note that the name hydrogenases best reflects the reverse reaction when a molecule (P) in an oxidized state gets reduced (to S) and H2 gets oxided to H+.
Crystal structures of hydrogenases show them to be unique among metal-containing enzymes. They contain two metals bonded to each other. The metal centers can either be both iron or one each of iron and nickel. The ligands interacting with the metals are two classical metabolic poisons, carbon monoxide and cyanide. Passages for flow of electrons and H2 connect the buried metals and the remaining enzymes. The metals are also bound to sulfhydryl groups of cysteine side chains. It appears that two electrons are added to a single proton making a hydride anion which accepts a proton to form H2. In the two Fe hydrogenases, the geometry of the coordinating ligands distorts the bond between the two iron centers, leading to irons with different oxidation numbers. Electrons appear to flow from one center to the other, as does carbon monoxide as well. Ultimately, hydrogenases or small inorganic mimetics of the active site could be coated on electrodes and used to general H2 when placed in water in electrolytic experiments.
![]() Photosynthesis
on the Web - 2002
Photosynthesis
on the Web - 2002
D10. Links and References
![]() Photosynthesis
on the Web - 2002
Photosynthesis
on the Web - 2002
- Current Opinion in Chemical Biology (2012) 16, 11–18. DOI 10.1016/j.cbpa.2012.03.003
- Nature (2011) 473, 55,-61.
- Journal of Biophysical Chemistry (2012) 3, 111-126. doi:10.4236/jbpc.2012.32013
- Coordination Chemistry Reviews (2012) 256, 2445– 2452
- Sessions, A. et al. The Continuing Puzzle of the Great Oxidation Event. Currrent Biology, 19, R567–R574 (2009)
- Alper, J. Water Splitting Goes Au Naturel. Science, 299, pg 1686 (2003)
- Demming-Adams, B. & Adams, W. Antioxidants in photosynthesis and human nutrition. Science. 298, pg 2149 (2002)
- Jordan et al. 3D Structure of Cyanobacterial Photosystem I (shows how chlorophylls use solar energy to transport electrons) Nature. 411. pg 896, 909 ((2001)
- Balabin and Onuchic. Exploiting thermal motion in electron transfer (combined molecular dynamics, quantum mechanics to study e transfer in photosynthetic reaction center) Science. 290. pg 61, 114 (2000)
- Dismukes. Photosynthesis: Splitting Water. Science. 292. pg 447 (2001)
Navigation
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8D: The LIght Reaction of Photosynthesis

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.