Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
B: PROBING COMPOSITION, SEQUENCE, AND CONFORMATION
BIOCHEMISTRY - DR. JAKUBOWSKI
2/28/16
|
Learning Goals/Objectives for Chapter 2B: After class and this reading, students will be able to describe in general terms the procedures and chemical steps in the determinations of the following for proteins:
|
B5. Analysis of Protein Tertiary Structure
Clearly, the highest resolution understanding of protein structure requires a solution to the 3D structure of the protein. Once that is determined, it is easy to devise computer programs which will determine what part of the structure is in secondary structure. Three methods are presently useful to determine the 3D structures of proteins.
A. X-Ray crystallography
If crystals of the protein can be made, traditional x-ray crystallographic techniques can be used to solve the structure. X-rays irradiate a crystal, which scatters the x-rays, leading to constructive/destructive interference patterns. Using appropriate math, the interference pattern can be reconverted into the actual structure of the protein. Check out a fun example showing such a reconstruction of the Parthenon from its diffraction pattern!
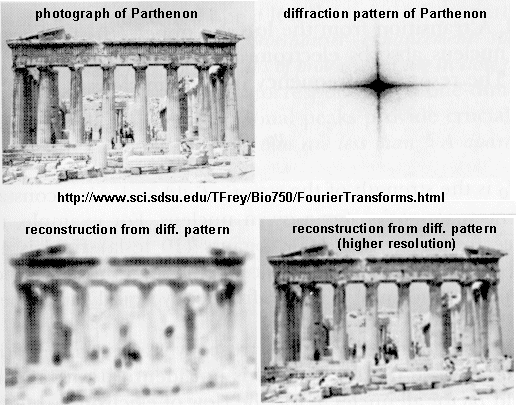
These structures are not solution structures, but overwhelming evidence suggest that they do represent the solution structure. For instance, x-ray structure contain many water molecules which interact with each other and the protein, a finding expected for a structure that represents the solution structure of the protein. In addition, substrates and inhibitors can be infused into the crystal and bind with the protein, suggesting again a native-like structure for the protein in the crystal.
B. NMR
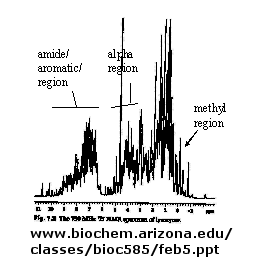
There are many protons in proteins which give a proton NMR spectra. The problem is one of assignment, since there are so many. Nuclei in different environments absorb energy at different resonant frequencies. When a proton spin flips, it goes to a higher energy state. It will return to the equilibrium state with some time delay.
Figure: 1D NMR spectra of a protein
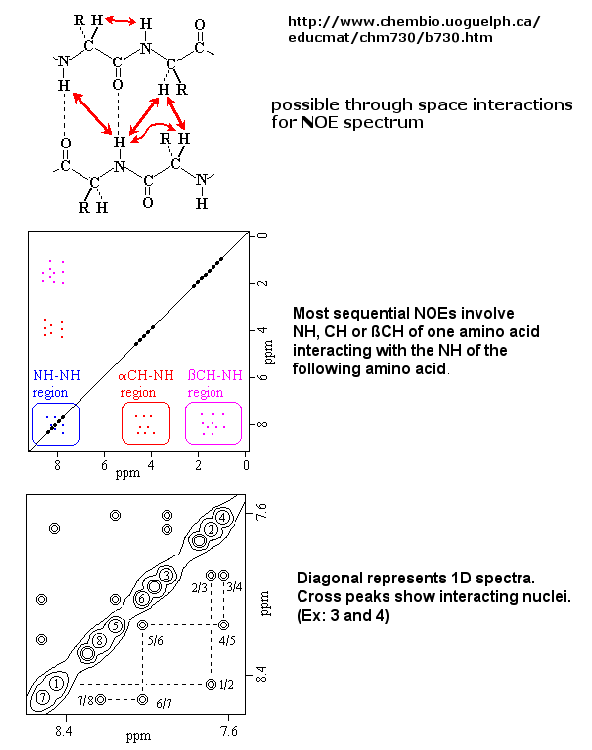
If an unexcited proton is proximal in space, the magnetization can be transferred to the unexcited proton. This interaction is inversely proportional to the 6th power of the distance between them and is the basis of the Nuclear Overhauser Effect (NOE). A 2D NOE spectra shows peaks off the diagonal that are correlated, indicated that they are close in 3D space.
Figure: 2D NOSEY spectra of a protein
Multi-dimensional techniques, obtained when isotopes of N(15) and C(13) are present in the protein, can be used to actually obtain a 3D solution of a small protein. NMR and X-ray structure of the protein are almost superimposable.
Figure: NMR Structure of Proteins
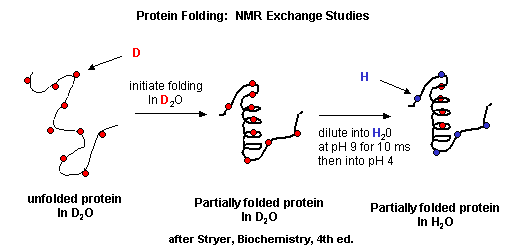
- Start2Fold: Database of hydrogen/deuterium exchange data on protein folding and stability Great link to visualize D2O folding data. TBA
-
 basic
NMR review
basic
NMR review -
 protein
NMR
protein
NMR -
 Structure
Determination of Proteins with NMR Spectroscopy. (including 1, 2,
and 3 D)
Structure
Determination of Proteins with NMR Spectroscopy. (including 1, 2,
and 3 D) -
 2D
NMR
2D
NMR
C. Homology Modeling
The structure of unknown proteins can be modeled theoretically if they have extensive sequence homology to another protein whose structure is known.
- homology modeling : general description
- homology modeling for beginners: a course
- an automative comparative protein modeling server - from the The ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB)
- comparative protein modeling from ExPASy.
Of course, to characterize proteins rigorously, they must be purified from a solution containing many proteins. We will cover these techniques in the laboratory. Here is a quick summary of some of them:
Navigation
Return to Chapter 2B: Probing Composition, Sequence and Conformation Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2B: Probing Composition, Sequence and Conformation

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.