Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 2 - PROTEIN STRUCTURE
C: UNDERSTANDING PROTEIN CONFORMATION
BIOCHEMISTRY - DR. JAKUBOWSKI
3/4/16
|
Learning Goals/Objectives for Chapter 2C: After class and this reading, students will be able to
|
C2. Secondary Structure
Secondary structures are those repetitive structures involving H bond between amide H and carbonyl O in- the main chain. These include alpha helices, beta strands (sheets) and reverse turns..
Alpha Structure
Figure: Right Handed Alpha helices - image made with VMD
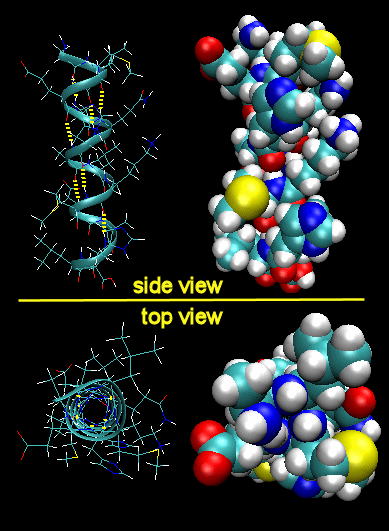
These helices are formed when the carbonyl O of the i th amino acid H bonds to the amide H of the i th +4 aa (4 amino acids away). The phi/psi angles for those amino acids in the alpha helix are - 57,-47, which emphasizes the regular repeating nature of the structure. It can also be characterized by n (the number of amino acid units/turn = 3.6) and pitch (the helix rise/turn = 5.4 angstroms). Some facts:
- the alpha helix is more compact than the fully extended polypeptide chain with phi/psi angles of 180o
- in proteins, the average number of amino acids in a helix is 11, which gives 3 turns.
- the left-handed alpha helix, although allowed from inspections of a Ramachandran plot, is never observed, since the side chains are too close to the backbone.
- the core of the helix is packed tightly. There are not holes or pores in the helix.
- All the R-groups extend backward and away from the helix axis.
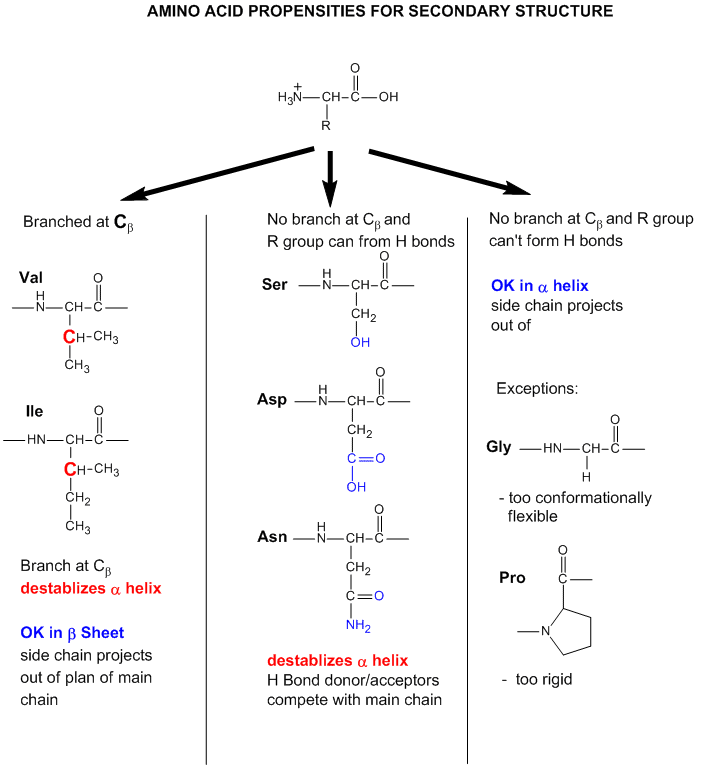
- Some amino acids are more commonly found in alpha helices than other. Amino acids can be divided into two kinds, those with branches at the beta C and those with none. Consider first those that aren't branched . Gly is too conformationally flexible to be found with high frequency in alpha helices, while Pro is too rigid. The amino acids with side chains that can H-bond (Ser, Asp, and Asn) and aren't too long appear to act as competitors of main chain H bond donor and acceptors, and destabilize alpha helices. The rest with no branches at the beta C can form helices. Those with branches at the beta carbon (Val, Ile) destabilize the alpha helix due to steric interactions of the bulky side chains with the helix backbone. (Remember left-handed alpha helices are not found in nature for similar reasons.) Summary of amino acids propensities for alpha helices (and beta structure as well)
- alpha keratins, the major component of hair, skin, fur, beaks, and fingernails, are almost all alpha helix.
![]() Jmol:
Updated An isolated helix from an Antifreeze Protein
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated An isolated helix from an Antifreeze Protein
Jmol14 (Java) |
JSMol (HTML5)
Note: There are other kinds of helices that can occur. These include a 310 helix and a p helix, which are stabilized by H-bonds between the amide NH and carbonyl O of residues (i, i+3) and (i, i+5), respectively. Likewise, they have 3 and 4.3 residues/turn, respectively, and a rise per residue of 6 and 4.7 angstrom, respectively. These structures are much rarer than right handed alpha helices.
| Helix Type | H bond btw ith and ith+X AA, where X = | Residue/turn | Rise (Angstrom)/turn |
| 310 | 3 | 3 | 6 |
| a | 4 | 3.6 | 5.4 |
| p | 5 | 4.3 | 4.7 |
Beta Structure
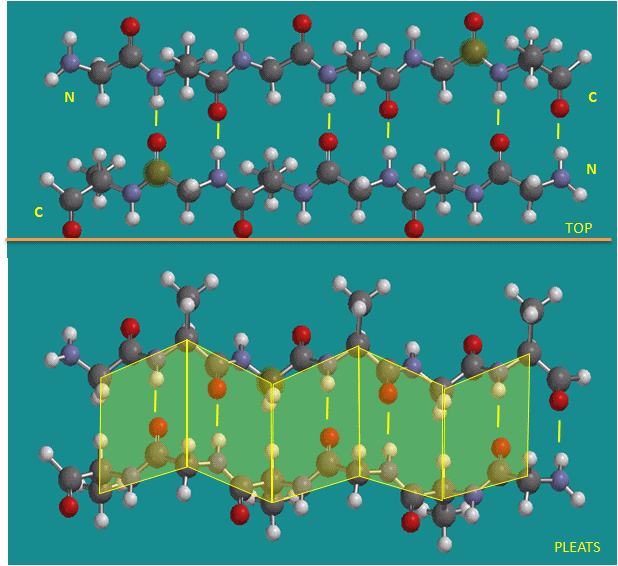
Beta Structure: Parallel and antiparallel beta strands are much more extended than alpha helices (phi/psi of -57,-47) but not as extended as a fully extended polypeptide chain (with phi/psi angles of +/- 180). The beta sheets are not quit so extended (parallel -119, +113 ; antiparallel, -139, +135), and can be envisioned as rippled sheets. They can be visualized by laying thin, pleated strips of paper side by side to make a "pleated sheet" of paper. Each strip of paper can be pictured as a single peptide strand in which the peptide backbone makes a zig-zag along the strip, with the alpha carbons lying at the folds of the pleats. Each single strand of the beta-sheet can be pictured as a twofold helix, i.e. a helix with 2 residues/turn. The arrangement of each successive peptide plane is pleated due to the tetrahedral nature of the alpha C. The H bonds are interstrand, not intrastrand as in the alpha helix.
Figure: Parallel beta strands (image made with Spartan)
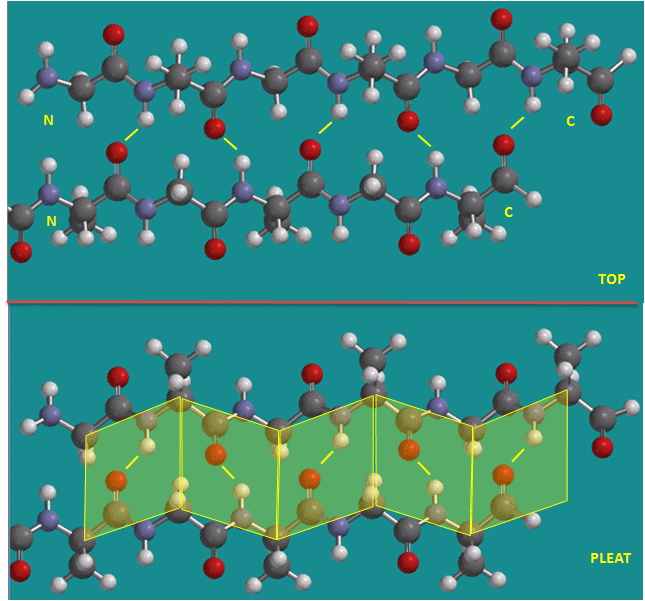
Figure: Antiparallel beta strands (image made with Spartan)
Note: Consider a strand as a continuous and contiguous polypeptide backbone propagating in one direction. Hence, using this definition, a helix consist of a single strand, and all the H-bonds are within the strand (or intrastrand). A beta sheet would then consist of multiple strands, since each "strand" is separated from other "strands" by an intervening contiguous stretch of amino acid which bends within the protein in a way which allows the next section of the peptide backbone, the next "strand", to H-bond with the first "strand". But remember, even in this case, all the H-bonds holding the alpha and beta structure together are intramolecular.
In a parallel beta sheet structure, the optimal H bond pattern leads to a less extended structure (phi/psi of -119, +113) than the optimal arrangement of the H bonds in the antiparallel structure (phi/psi of -139, +135). Also the H bonds in the parallel sheet are bent significantly. (i.e. the carbonyl O on one strand is not exactly opposite the amide H on the adjacent strand, as it is in the antiparallel sheet.) Hence antiparallel beta strands are presumably more stable, even though both are abundantly found in nature. Short parallel beta sheets of 4 strands or less are not common, which might reflect their lower stability.
The side chains in the beta sheet are normal to the plane of the sheet, extending out from the plane on alternating sides. Parallel sheets characteristically distribute hydrophobic side chains on both side of the sheet, while antiparallel sheets are usually arranged with all the hydrophobic residues on one side. This requires an alternation of hydrophilic and hydrophobic side chains in the primary sequence. Antiparallel sheets are found in silk with the sheets running parallel to the silk fibers. The following repeat is found in the primary sequence: (Ser-Gly-Ala-Gly)n), with Gly pointing out from one face, and Ser or Ala from the other.
![]()
![]() Jmol:
Silk
Jmol:
Silk
Beta strands have a tendency to twist in the right hand direction. This leads to important consequences in how the beta strands are connected. Parallel strands can form twisted sheets or saddles as well as beta barrels.
Figure: Twisted Beta Sheet/Saddle (image made with VMD)
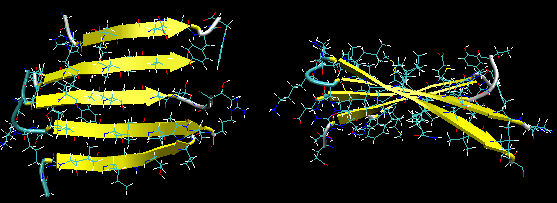
Figure: Beta Barrel (image made with VMD)

- in parallel strands, right handed connectivity is common.
- in a protein with parallel strand in register, and given the inherent twist in the stands, the strands arrange in a way to have the H bonds stretched equally at the ends of the chains, giving rise to a twisted saddle shape (top structure above).
![]() Jmol:
Updated Twisted beta sheet from Arabinose Binding Protein
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Twisted beta sheet from Arabinose Binding Protein
Jmol14 (Java) |
JSMol (HTML5)
- in a protein with parallel strand out of register, and given the inherent twist in the stands, the strands arrange in a way to have the H bonds stretched equally at the ends of the chains, giving rise to a beta barrel (bottom structure above).
![]() Jmol:
Beta barrel from triose phosphate isomerase
Jmol:
Beta barrel from triose phosphate isomerase
Reverse Turns: About 50% of the amino acids in a globular protein are in regular secondary structure (alpha or beta). The remaining amino acids are not less ordered, just less regular. An additional example of secondary structures is reverse turns (or beta-bends or beta turns). Reverse turns often connect successive antiparallel beta strands and are then called beta hairpins.
Figure: Reverse Turns (image made with VMD)
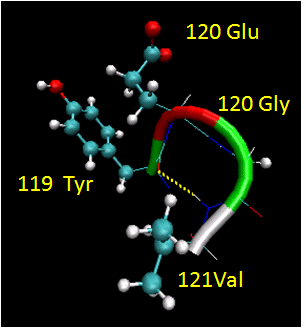
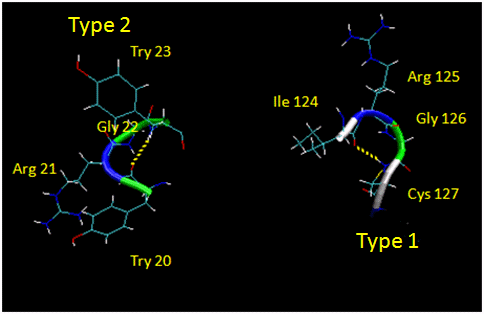
They are almost always at the surface, and consist of 4 amino acids. There are two types. (I - f2 = -60, y2=-30; f3 = -90, y3 = 0; II - f2 = -60, y2=120; f3 = 90, y3 = 0 ) Residue 2 of both is often Pro. (Why?) Both have an H bond between the carbonyl O of the i th a.a and the amide H of the i th+3 aa (three amino acids away). In the type 2, the O of residue 2 crowds the beta C of residue 3, so aa2 is usually Gly. Why? Those amino acids which destabilize alpha helices are often found in beta sheets, since the side chains project out of the plan which holds the main chain.
Figure: Type I and Type II Reverse Turns - from hen egg white lysozyme (image make with VMD)
![]() Jmol:
Updated Reverse Turn: Trypsin Inhibitor
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Reverse Turn: Trypsin Inhibitor
Jmol14 (Java) |
JSMol (HTML5)
(Notice the tightness of the reverse
turn and the presence of Pro and
Gly.)
Figure: Why do amino acid propensities for secondary structure differ?
-
 Secondary
Structure and Backbone Conformation from
ExPASy.(concentrate on second part on secondary structure)
Secondary
Structure and Backbone Conformation from
ExPASy.(concentrate on second part on secondary structure)
Navigation
Return to Chapter 2C: Understanding Protein Conformation Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 2C: Understanding Protein Conformation

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.