Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 9 - SIGNAL TRANSDUCTION
C: SIGNALING PROTEINS
BIOCHEMISTRY - DR. JAKUBOWSKI
04/16/16
|
Learning Goals/Objectives for Chapter 9C:
|
Estonian Translation √ by Anna Galovich
C10. AMP dependent protein kinase (AMPK)
AMP Kinase is one of the cell's major fuel sensors and also in mammals responds systemically to hormone and nutrient levels. The enzyme is a heterotrimeric protein consisting of an alpha (catalytic), beta (regulatory) and gamma (regulatory) subunit that binds AMP, ADP, and ATP. Cellular ATP levels are determined in part by the enzyme adenylate kinase which helps interconvert adenine nucleotide (AXPs) as shown in the following equilibrium:
Adenylate Kinase: ADP + ADP <==> ATP + AMP, Keq = 0.44
In a red blood cells, the concentrations of ATP, ADP and AMP are approximately 1850 uM, 145 uM and 5 uM. Even in cells that use lots of ATP (muscle for example), ATP never falls by much. Using the values above and simple general chemistry, an 8% drop in ATP would lead, through the action of adenylate kinase, to an ATP concentration of about 1710 uM and an AMP concentration of 20 uM. This value for AMP is still much lower than ADP and ATP. However, this change represents a 4 fold increase in AMP which, even with the low actual concentration of AMP, leads to activation of AMPK.
Another "normalized" indicator of cell energy status (or "charge") is the Energy Charge, EC. It is defined by an equation that give an EC value from 0-1 where 0 indicates that all AXPs are in the AMP form, and 1 where only ATP is present. The numerator of the equation of EC below represents the number of moles of phosphoanhydride linkages in the AXP pool (two for each ATP and one for ADP) and the denominator the number of moles of AXPs (mass balace). The 1/2 term allows the bracketed term to equal 1 when only ATP exists and 0 when only AMP exists. The EC values of cells is regulated to remain around 0.85.
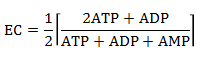
How AMPK detects this exponential but still small molar increase in AMP is interesting, especially given the much higher concentrations of ADP and ATP. AMPK contains four binding sites that can bind AMP, ADP, and ATP (AXPs) in the regulatory subunit (gamma). This is in addition to the binding of ATP and ADP at the active site in the catalytic subunit (alpha). What binds depends on the Kd for binding of different AXPs as well as their concentrations. Bound AMP seems to have three effects on AMPK. When bound to the gamma subunit, AMP
-
increases phosphorylation of Thr 172 in an "activation" loop in the catalytic alpha subunit by an upstream kinase which increases the kinase activity of AMPK by 100-200 fold. Phosphorylation is essential for the activity of the enzyme;
-
inhibits dephosphorylation of Thr 172 which is perhaps the major way that AMP enhances the kinase activity of the catalytic subunit. ADP binding also inhibits dephosphorylation as shown by studies that show that the binding of ADP and the dephosphorylation of the phospho-AMPK have the same ADP concentration dependency;
-
allosterically activates by ten-fold the kinase activity of the catalytic alpha subunit (a secondary effect). ADP has no such effect.
These effects are altered by the markedly higher concentrations of ATP which counteracts all these effects, enhancing the Energy Charge sensor activity of this enzyme.
The gamma regulatory subunit has 4 binding sites for AXP. Crystal structures show site 2 is empty, site 4 is always bound to AMP and sites 1 and 3 can bind AMP, ADP, or ATP (Xiao et al, 2011). Site 1, which mediates the allosteric effects on AMPK binds all AXPs with similar affinity. This appears paradoxical since given the high energy charge, one would expect ATP and ADP to outcompete AMP for binding. However, it was found that the Mg2+ -ATP complex has marked lower affinity for the site, allowing both AMP and ADP, which under cellular conditions are mostly not bound to Mg2+ while ATP is, to outcompete Mg2+ -ATP for binding. Site 3 binds AMP and ADP with a 30-fold lower affinity but on binding protects p-AMPK from dephosphorylation of Thr 172.
![]() Updated
AMP-dependent Protein Kinase (AMPK)
Jmol14 (Java) |
JSMol (HTML5)
Updated
AMP-dependent Protein Kinase (AMPK)
Jmol14 (Java) |
JSMol (HTML5)
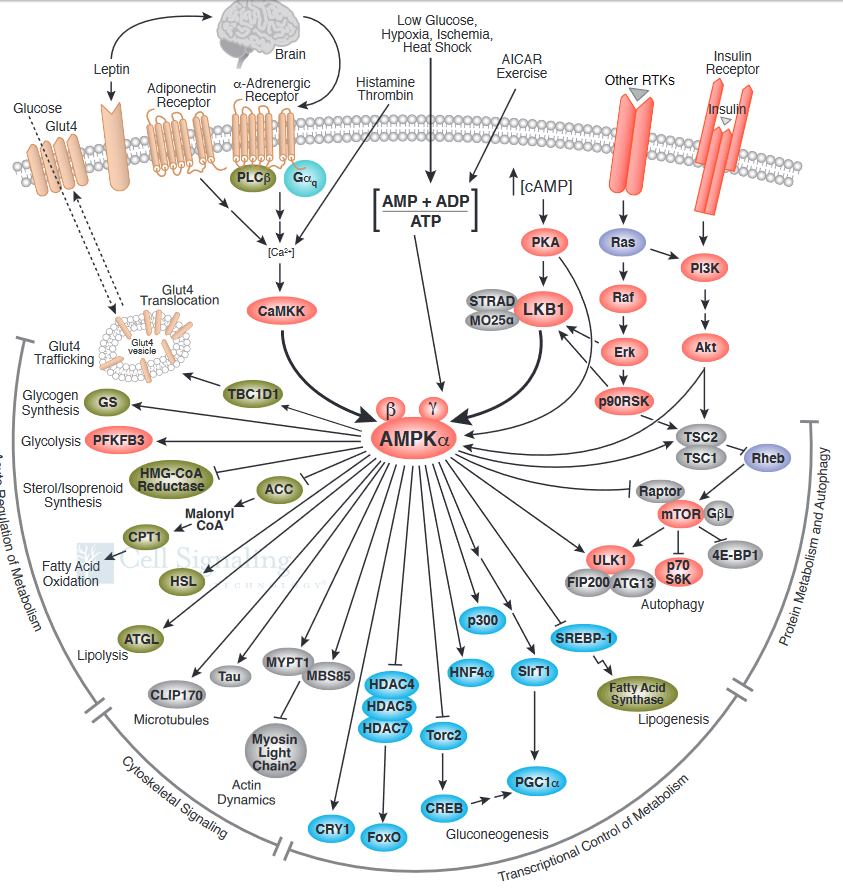
What effect does activated AMPK have on the cell? Active AMPK has an amazing number of effects (see figure below). It activates liver glycolysis (by phosphorylating phosphofructokinase 2 which forms F2,6-BP, an activator of PFK) and inhibits by phosphorylation enzymes involved in fatty acid synthesis (acetyl-CoA carboxylase), glycogen synthesis (glycogen synthase) and cholesterol synthesis (HMG-CoA reductase). Yeast AMPK has recently been shown to be also controlled by acetylation of the equivalent beta subunit (Sip2). Acetylation increases its interaction with the alpha catalytic subunit (Snf1) which decreases its kinase activity. This decreases phosphorylation of downstream kinases (including an analog of Akt1 called Sch9) which slows growth and increases longevity. Normal aging is associated with decreased acetylation of Sip2.
The figure below shows a more complete pathway of activation, regulation, and activity of AMPK. The illustration is used with courtesy of Cell Signaling Technologies (www.cellsignal.com)
It also inhibits a master regulator of protein, lipid and nucleic acids synthesis, mTOR (also shown below). Synthesis of these molecules is necessary for cell growth and proliferation, two activities that cells do not engage in when AMP levels are high, which signifies an energy depleted state.. A whole chapter section, Chapter 9D: Nutrient Signaling, is devoted to the involvement of mTOR in these processes.
Navigation
Return to Chapter 9C. Signaling Proteins Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 9C: Signaling Proteins

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.