Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 9 - SIGNAL TRANSDUCTION
B: NEURAL SIGNALING
BIOCHEMISTRY - DR. JAKUBOWSKI
06/10/14
|
Learning Goals/Objectives for Chapter 9B:
|
B1. Introduction to Neural Signaling
Neurochemistry is one of the most explosive areas of biological research. Scientists are now starting to unravel the molecular bases for memory, cognition, emotion, and behavior. The next decades will bring truly revolutionary understanding of brain chemistry and along with it the potential to alter human mood, memory, and to treat mental illness such as schizophrenia much more effectively. The human brain, with about 100 billion neurons (each which can form connections - synapses - with 1000 to 10,000 other neurons ) and associated glial cells (10-50 times the number of neurons) can be considered one of the most complex structures in the universe. This section will explore the biology and chemistry of neurons.
We will discuss two kinds of neurons - those that interact with muscles at the neuromuscular junction and those that interact with other neurons in the central nervous system. Neurons consist of a single, nucleated cell body with multiple signal-receiving outgrowths (dendrites) and multiple-signal sending outgrowths (axons) which end in a terminal button. These interact through the synapse with dendrites on other neurons.
Figure: Neurons
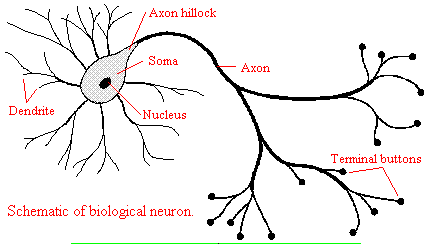
A presynaptic neuron can stimulate an adjacent postsynaptic neuron by releasing a neurotransmitter into the synapse between the cells, which binds to a receptor in the membrane of the post-synapatic cell, stimulating the cell.
Figure: neurotransmitter into the synapse between the cells
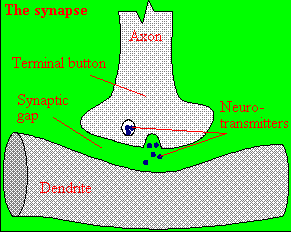
We will discuss the events which cause the post-synaptic cell to "fire", but we will not discuss the immediate events which lead to the release of neurotransmitter by the presynaptic neuron.
Neurons (as do all cells) have a transmembrane potential across the membrane. Transient changes in the membrane potential are associated with neuron activation or inhibition. This arises in part due to the imbalance of ions across the membrane which was established by the Na+-K+-ATPase in the membrane. This electrogenic antiporter transfers 3 Na ions out of the cytoplasm for every 2 K ions it transports in. Likewise Cl- has a much higher level outside the cell. Membrane potentials are determined not only by the size of the ion gradients across the membrane, but also the differential permeability of membranes to ions. As we saw previously, synthetic bilayer membranes are not very permeable to ions .
Ion permeability of phosphatidyl serine vesicles
| ION | PERMEABILITY (cm/s) |
| sodium | <1.6 x 10-13 (lowest) |
| potassium | <9 x 10-13 |
| chloride | <1.5 x 10-11 (highest) |
Sodium would be expected to have a lower permeability than potassium since it has a higher charge density. It also is largest in effective size due to the larger hydration sphere around the ion arising from its higher charge density. Chloride would be expected to have the highest permeability since it has the lowest charge density (due to repulsion of the electron cloud in the negative ion). Intracellular charged proteins (which are mostly negative) are not permeable and help in creating the negative charge imbalance across the membrane.
Much work has been done on the giant axon of the squid, which has uniquely high intracellular potassium (400 mM compared to 20 mM outside) and high extracellular fluid sodium (440 mM vs, 50 mM inside) and chloride (560 mM vs 52 mM inside). Mammalian cell concentrations are much lower, but the relative size of the gradient is about the same.
Typical ion concentrations and permeabilities for mammalian membranes.
| Ion | Cell (mM) | Blood (mM) | Permeability (cm/s) |
| potassium | 140 | 5 | 5 x 10-7 |
| sodium | 5-15 | 145 | 5 x10-9 |
| chloride | 4 | 110 | 1 x 10-8 |
| X- (neg. macromol.) | 138 | 9 | 0 |
How can we account for the markedly greater permeabilities of ions (1000x to 1,000,000 x) in mammalian cell membranes compared to synthetic lipid vesicles? Previously, we showed that glucose has a greater permeability through red blood cell membranes than through synthetic liposomes because of a membrane receptor that allows facilitated diffusion across the membrane and down a concentration gradient. The same thing is true of ion permeabilites in intact biologicial membranes. These membranes have several types of selective ion channels (nongated - always open, and gated - open only after specific conformational changes). The nongated channels dramatically increase the permeability of membranes to ions, as the glucose transport protein increased the permeabilty to glucose. It turns out that this differential permeability contributes to the transmembrane potential. Ion channels in nerve and muscle can move ions across the membrane at a rate up to 109/s, which is comparable to kcat for the best enzymes.
If we envision channels as pores, how can we account for the selectivity of the channel to specific ions. A larger pore should admit any ion less than a maximal size for the pore, so it is hard to image the nature of the selectivity filter. Because of this, many people discounted the ideas of channels in favor of a transporter, which would bind the ion selectivity and then, through conformational changes, move the ion across (much like the Na/K ATPase we discussed in the previous guide). This model could not, however, account for the incredible rates of ion flux across the membrane. Selectivity can be accounted for by a channel that contains a narrow opening that acts as an ion sieve. The ion looses most of its hydration sphere and would form specific interactions with amino acid side chains in the pore region. Such an interaction would be transient and not too tight since the ion must pass through the membrane. As we will see later, these ion channels:
- pass ions down a concentration gradient in a thermodynamically favorable process
- are specific for certain ions (although a few are less selective and will pass Na, K, Ca, and Mg ions)
- allow ion flow through either ungated or gated channels.,
- saturate with increasing ion concentration (even though as concentration increases, the ions have a greater thermodynamic drive to pass through the channel). This is consistent with binding of the ion at a selectivity filter in the narrow part of the pore. The Kd for the interaction is usually in the mM range and indicates weak binding with large dissociation rate constants (koff).
B2. Transmembrane Potentials
Several questions arise about the distribution of ions and the magnitude of the transmembrane potential.
- How are the ion gradients established?
- How does the transmembrane ion distribution contribute to the membrane potential?
- How can the resting electrochemical potential and the ion distribution be maintained?
The answer to these questions will be
illustrated using studies on two types of brain cells, glial cells (which
function as protectors, scavengers, and feeder for brain neurons) and
neurons. Both types of cells have transmembrane potentials.
Glial Cells
1. The transmembrane ion gradients for ions can
be established by different mechanisms. One uses ion-specific ATPases
(P-type ion transporters), such as we discussed with the Na/K ATPase. This
transporter ejects 3 sodium ions from the inside of the cell for every 2
potassium ions it transports in, all against a concentration gradient.
Since it is an electrogenic antiporter, it helps generate the potential.
Additionally, specific ion channels also contribute (as described below) to
the transmembrane gradients and potentials.
2. The harder question
is how the ion distribution contribute to the membrane potential. Two
things must occur for a membrane potential to exist: First, there must
be a concentration gradient of charged ions (for example, sodium, potassium,
or chloride) across the membrane. Second, the membrane must be
differentially permeable to different ions. If the membrane were
completely impermeable to ions, then no movement of ions across the membrane
could occur, and no membrane potential would arise. If, however,
membranes are differentially permeable to the ions, an electrical potential
across the membrane can arise. Remember, synthetic bilayers are
quite impermeable to ions, given the hydrophobicity of the internal part of
the bilayer. Likewise it is quite impermeable to glucose. It turn out that
glial cells appear to have only a non-gated potassium channel, which allows
the outward flow of potassium ions down the concentration gradient. The
inside will then have a net negative charge since impermeable anions remain.
The chemical potential gradient causes this outward flow of potassium ions.
As more ions leave, the inside gets more negative, and a transmembrane
potential develops which resists further efflux of potassium. Eventually
they balance, and the net efflux of potassium stops. The resting
transmembrane potential reaches -75 mV which is exactly the value obtained
from the equations we will derive below. Since glial cells appear to only
express a nongated potassium channel, their resting potential is equal to
the potassium equilibrium potential.
Figure: Visualizing the transmembrane potential in K+ loaded vesicles + a nongated K+ channel.

(Note: above figure has mistake. Cl- ion , assuming that is not permeable, will not change across the membrane at equilibrium.)
B3. Thermodynamics of Membrane Potential
How can this equilibrium potential be calculated? The total thermodynamic driving force,ΔGtot , for an ion X moving across the membrane (from inside to outside) of transmembrane potential ΔΨ is the sum of the ΔGs arising from concentration gradients and from the electrical potential:
DGtot = ΔGion grad + ΔGelect pot
The ΔGelect pot can be determine from the simple general chemistry equation: ΔGelect pot= -nFE = -nFΔΨ
where F, the Faraday constant (the electric charge on one mol of electric charge = 23,000 cal/V.mol), n (or z) is the number of moles of charge transferred per mol of ions moving (+ 1 for Na and K ions) and ΔΨ ( or E) is the transmembrane electrical potential in volts. Hence for the movement of an ion from inside to outside the cell,
DGtot = [Gout - Gin] - zFDY
This shows that the total driving force for ion movement consists of a chemical potential (first term) plus an electrical potential (second term). The equation can be expanded and rearranged as follows, where Xin and Xout are the concentrations of ion X inside and outside the cell, respectively:
DGx = (ΔGxo + RTlnXout) - (ΔGxo + RTlnXin) - zFDY
DGx = RTlnXout - RTlnXin - zFDY
1) ΔGx = RTln(Xout/Xin) - zFDY
2) ΔGx = 2.303RTlog(Xout/Xin) - zFDY
Consider K+, where Xout/Xin is about 1/20 = 0.05. (The table above suggests that it is more like 1/35, but some texts use the value of 20.) What would the transmembrane potential have to be so that no driving force would exist for K ions to move across the membrane (i.e. under equilibrium conditions)? Under these conditions, ΔGx = 0, so
3). 2.303RT log(Xout/Xin ) = zFΔΨeq or
4. ΔΨeq = (2.303RT/zF) log(Xout/Xin ) or
5) ΔΨeq = (0.0615/z) log(Xout/Xin) at 310K or 37oC) or
6) ΔΨeq = (0.0591/z) log(Xout/Xin ) at 298K or 25oC
Equations 4-6 are versions of he Nernst equation which you remember from General and Analytical Chemistry.For K+ ions, ΔΨ is about -0.075 V or -75 mV (assuming 37oC = 310K, R = 1.98 cal/(K.mol), and z = +1). That is, since the inside is more negative than the outside, then K ions would tend to stay inside even if the chemical potential of K ions is greater on the inside. A similar calculation for Na ions, with a Xout/Xin of about 12, gives an equilibrium Na ion potential of about +55 mV. Why is the actual transmembrane potential close to the equilibrium potential of K ions and not that of Na ions? The reason has to do with the fact that the permeability of K+ ions is 100 fold greater than of Na ions. Hence it is the K ions which mostly determine the cell resting potential. (Remember glial cells have only a nongated K channel.)
In general, the ΔΨeq is determined by more than one ion (usually Na and K), so the actual ΔΨeq is between the ΔΨeq for each ion alone. A more general equation can be derived which shows that the ΔΨeq is determined not only by the concentrations but also the permeability of these ions. (In a liposome made with encapsulated KCl at a higher concentration than the outside concentration, and with both sides being electrically neutral, if no ions could flow across the membrane, no membrane potential would arise.) The equation that considers both concentration AND permeability effects is the Goldman equation and is given by:
DYeq = (RT/F) ln {[PKK+out + PNaNa+out + PClCl-in] / [PKK+in + PNaNa+in+ PClCl-out}
DYeq = 2.303 (RT/F) log {[PKK+out + PNaNa+out + PClCl-in] / [PKK+in + PNaNa+in + PClCl-out]}
DYeq (in volts) = 0.059 log {[PKK+out + PNaNa+out + PClCl-in] / [PKK+in + PNaNa+in + PClCl-out]}
DYeq (in mV) ~ 59 log {[PKK+out + PNaNa+out + PClCl-in] / [PKK+in + PNaNa+in + PClCl-out]}
where PX is the permeability of ion X. This treatment recognizes the importance of thermodynamics (chemical and electrical potentials) and kinetics (permeability coefficients). Remember this potential would not be possible if the ion gradient was not maintained by Na/K ATPase.
To see the effects on the transmembrane voltage use the Goldman Equation Calculators below and plug in the following generally realistic values for concentrations and permeabilities of ions in neurons:
| ion | inside (mM) | outside (mM) | permeability coeff (cm/s) |
| K+ | 140 | 5 | 1 x 10-8 |
| Na+ | 12 | 145 | 1 x 10-10 |
| Cl- | 3 | 145 | 1 x 10-10 |
Then increase the permeability of Na ions and watch the effect on the transmembrane voltage. Reset the Na ion permeabilty and change that for chloride ion.
-
 Goldman
Equation simulator
Goldman
Equation simulator -
 Goldman
Equation Calculator (not updated with new Java, 3/18/14)
Goldman
Equation Calculator (not updated with new Java, 3/18/14) -
 Applets
for action potentials (not updated with new Java, 3/18/14)
Applets
for action potentials (not updated with new Java, 3/18/14)
Fluorophores that insert into membranes and whose flourescence changes with transmembrane potential have been developed. An example is di-4-ANEPPS, a AminoNaphthylEthenylPyridinium dye, whose structure is shown below.
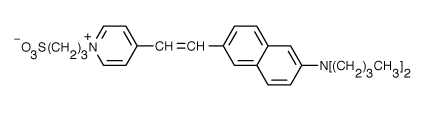
When bound to membranes, this probe has an excitation and emission maximum of 475 and 617 nm, respectively. When the membrane potential is made more negative (hyperpolarized), the fluorescence intensity (excitation 440 nm) decreases while the emission increases when excited at 530 nm.
In the 1940's, ways were developed to measure the actual transmembrane potential of cells. Varying the outside sodium and potassium concentrations would change the experimental transmembrane potential, as indicated by the Nernst equation. The experimental resting potentials of glial cells always matched the theoretical potassium equilibrium potentials, supporting the view that the transmembrane potential was associated only with open, nongated potasisum channels. This was not observed with neurons, suggesting that channels other than for potassium were open. Using radioactive tracers and potential measurements, it became clear that nerve cells were permeable not only to potassium, but also to sodium and chloride. How do these work in establishing the resting potential? Consider the simplest case when just potassium channels are present, along with an unequal distribution of other ions. Now add some sodium channels. Two forces act to drive sodium into the cell - the chemical potential since sodium is higher on the outside, and the electrical potential since the inside of the cell is negative. The equilibrium potential of a cell if it were only permeable to sodium is +55 mv, so there is a great electrical drive for sodium to enter through the nongated, open sodium pores we just added. As sodium enters, the cell starts to "depolarize" and have a more positive voltage. However, since in our example, there are many more open potassium channels, the resting potential deviates only a small amount from the potassium potential, since as the potential becomes more positive, more potassium flows out down the concentration gradient. Eventually the enhanced potassium efflux equals the sodium influx, and a new resting membrane potential of -60 mV is established, which is typical of neurons.
Now we can answer
question 3 above: How is the resting electrochemical potential and the
transmembrane ion distribution maintained? For a resting electrical
potential to be maintained once established, the rate of sodium influx must
equal the rate of potassium efflux through the open, non-gated channels in
order to maintain separation of charge (and the electrical potential) across
the membrane. But, if influx and efflux were allowed to continue, the actual
ion gradients (chemical potential) across the membranes would collapse. This
problem is solved by the Na/K ATPase. In the resting cells, the passive
fluxes of sodium and potassium ions are exactly balanced by the active
fluxes of these ions mediated by the Na/K ATPase. The cell is not really at
equilibrium but at a steady state.
B4. Neurotransmitter Activation of Neurons
What happens when a neurotransmitter binds to a receptor on the post-synaptic cell? We will study two examples. The first is the simplest: binding of the neurotransmitter acetylcholine, released by a motor neuron, to its receptor on muscle. This region is called the neuromuscular junction. Binding of acetylcholine will lead to a transient depolarization of the muscle cell. Next we will discuss the interaction of a neurotransmitter with a post-synaptic neuron in the central nervous system. This is a much more complex system. Their differences are described below:
In neurons interacting with muscles:
- Most muscle fibers are innervated by only one neuron - a motor neuron
- Neurotransmitter release at the neuromuscular junction leads only to muscle excitation, not inhibition.
- All fibers are excited by the same neurotransmitter - acetylcholine.
In the central nervous system, life is more complicated:
- Stimuli are received from hundreds to thousands of different neurons.
- Nerves receive both excitatory and inhibitory stimuli from neurotransmitters
- Different kinds of receptors are present to receive stimuli, which control the activity of different kinds of channels.
- The ion channels in neurons are gated by a variety of mechanisms in addition to changes in membrane potential, including gating by heat, cold, stretch, or covalent modification.
- Most nerve cells have a resting potential of about -65 mV compared to a -90 mV for a muscle cell.
What happens when a neurotransmitter binds to the receptor on the post-synaptic cell? A depolarization occurs (mediated by conformational changes in the transmitter-receptor complex), raising the membrane potential from the resting equilibrium level. What happens next depends on the identity of the post synaptic cell. In the muscle cell, the rising potential caused by binding of acetylcholine ultimately leads to muscle contraction by opening intracellular organelle membrane calcium channels. In a neuron, the rising potential triggers an action potential by opening voltage-gated sodium channels. The potential rises to about + 35 mV, but does not reach the Na ion equilibrium potential, because the high positive potential opens a voltage-gated potassium channel. The potential then falls until it reaches the K ion equilibrium potential where the cells is hyperpolarized. It slowly then relaxes back to the resting potential of -60 mV. This wave of changes in potential sweeps down the post-synaptic cell membrane and is the basis for the "firing" of the neuron.
Figure: DEPOLARIZATION OF TRANSMEMBRANE POTENTIAL
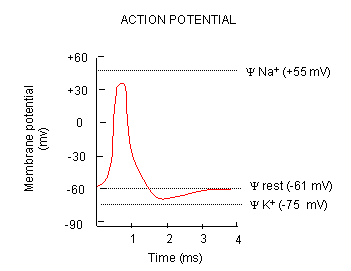
Figure: NA AND K PERMEABILITIES DURING DEPOLARIZATION
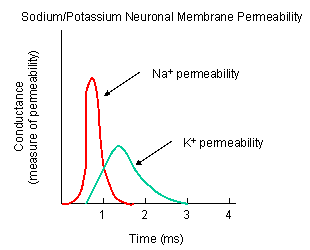
![]() Animation:
Voltage-Gated Channels and Action Potentials
Animation:
Voltage-Gated Channels and Action Potentials
![]() Animation:
Sodium Potassium ATPase
Animation:
Sodium Potassium ATPase
![]() Animation:
Channel Gating During An Action Potential
Animation:
Channel Gating During An Action Potential
![]() Animation:
Propagation of An Action Potential
Animation:
Propagation of An Action Potential
B5. Proteins of the Neural Synapse
We must now account for the rise and fall of the membrane potential to a variety of neurotransmitters, including the cholineric transmitters (ex. acetylcholine), catecholamines (dopamine, epinephrine, norepineprine), amino acid derivatives (ex. Glu, Asp, N-methyl-D-Asp, Gly, gamma-amino-butyric acid -GABA), and peptides (endorphins, encephalins). We will consider five membrane proteins:
Figure: five membrane proteins
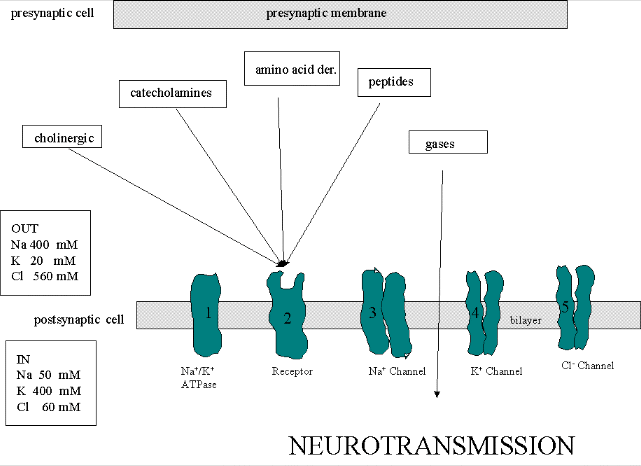
- Na+-K+-ATPase: We discussed this in the previous section on active transport. It transports both sodium and potassium ions against a concentration gradient using ATP as an energy source. The protein is a sodium dependent ATPase. Without this protein, the membrane potential could not be maintained since the sodium and potassium gradient would collapse. It also contributes to the potential since its is an electrogenic antiporter. (In addition, we have seen that ungated potassium and sodium channels are also present.)
- Neurotransmitter receptor: The receptors we will consider here are typically ligand-gated ion channels. Once ligand binds, a conformational change occurs in the protein, allowing a flow of ions down a concentration gradient. Depending on the nature of the ion, the channel either initiates depolarization (when Na+ enters from the outside and raises ΔΨ) or inhibits depolarization (when Cl- enters from the outside and lowers ΔΨ). When chloride channels open, they hyperpolarize the transmembrane potential. Stimulatory neurotransmitters (like glutamate) lead to depolarization of the membrane, while inhibitory neurotransmitters (like gamma-aminobutyric acid) lead to hyperpolarization of the membrane (make the potential more negative).
- Na+ channel (voltage-gated): When the ligand-gated channel depolarizes the membrane to some threshold value, sodium channels undergo a conformational change and open allowing Na+ ions to flood into the cell, raising the potential to a positive approx. 33 mV (a value lower than the the equilibrium sodium potential). This membrane protein is a voltage-gated channel, not a ligand gated one. Somehow, it senses a change in the transmembrane potential initiated by opening of the ligand-gated channel.
- K+
channel (voltage gated): When the membrane potential reaches around
+25 mV or so, the K+ channel, a voltage-gated membrane protein, alters
its conformation, allowing K+ efflux from the cells, lowering the
potential until it reaches the potassium equilibrium potential. It
slowly relaxes back to the cell resting potential of about -60 mV.
Recently, the crystal structure of the K+ channel from Streptomyces
lividans was determined (Science, 280, 69-77, 1998): "All K+ channels
show a selectivity sequence of K+, Rb+ > Cs+, whereas permeability for
the smallest alkali metal ions Na+ and Li+ is immeasurably low.
Potassium is at least 10,000 times more permeant than Na+, a feature
that is essential to the function of K+ channels.....Two aspects of ion
conduction by K+ channels have tantalized biophysicists for the past
quarter century. First, what is the chemical basis of the impressive
fidelity with which the channel distinguishes between K+ and Na+ ions,
which are featureless spheres of Pauling radius 1.33 � and 0.95 �,
respectively? Second, how can K+ channels be so highly selective and at
the same time, apparently paradoxically, exhibit a throughput rate
approaching the diffusion limit? The 104 margin by which K+ is selected
over Na+ implies strong energetic interactions between K+ ions and the
pore. And yet strong energetic interactions seem incongruent with
throughput rates up to 108 ions per second."
- Cl- channel: If these channels (typically ligand-gated) are open, they will hyperpolarize the cell and make it more difficult to fire.
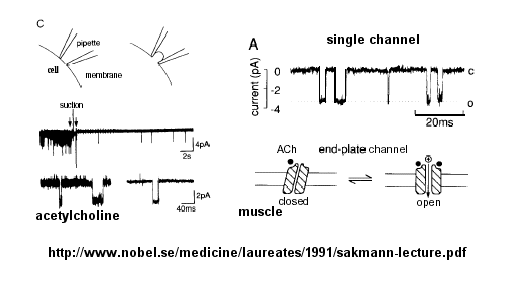
B6. Ligand-Gated Acetylcholine Receptor of the Neuromuscular Junction
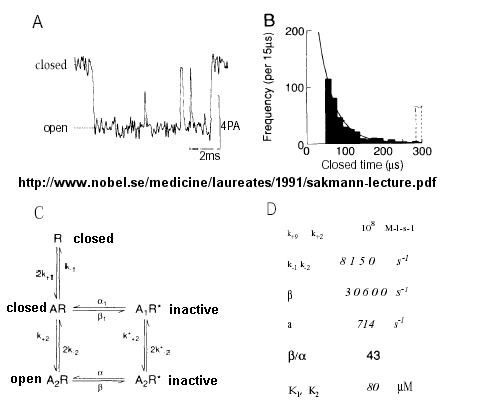
Kinetic modeling based on patch-clamp
recordings show that the protein can exists in multiple conformations,
including a closed (C), open (O), and inactive (I) forms. The binding of two
acetylcholine (A) molecules are required to open the channel. When two
are bound, the protein exist in two interconvertible forms: A2C <==> A2O.
Through another conformation change, the open form O can inactivate in a
slow step to A2I, where I is an inactive form.
The channel consists of
five helices which are constricted by hydrophobic side chains which point
inward into the pore. A rotation and sliding of the helices relative to each
other probably occurs when ligand binds the protein, allowing the channel
opening to increase. Rings of negatively changed side chains line the pore
at the top and bottom accounting for the selectivity of the channel for
positive ions.
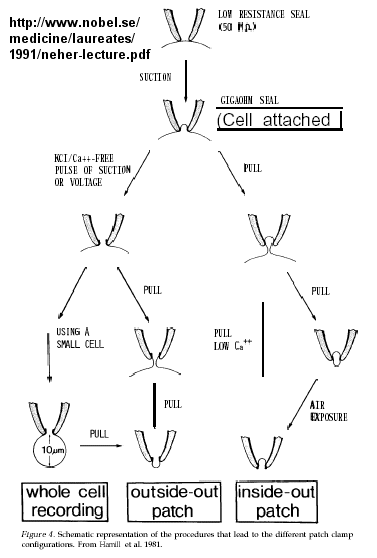
Much of our understanding of the mechanism of the acetylcholine channel opening comes from a patch clamp technique in which a micropipette is used to remove a small section of a membrane containing a channel.
Figure: patch clamp technique
Ion flow, measured as picoamp current, can be measured across the membrane patch. The opening and closing of single channels can be recorded using this technique, for which Nehert and Sakman won the Nobel Prize.
Figure: opening and closing of single channels
From this, they deduced a mechanism for gating of the acetylcholine channel.
Figure: mechanism for gating of the acetylcholine channel
In general, transmitter-gated channels are similar to the acetylcholine receptor. Mechanisms to generate an excitatory synaptic potential (with a transmitter-gated receptor/channel) are similar to that of generating an action potential (with voltage-gated channels) in that both involve movement of sodium and potassium ions across the membrane. They differ in several ways, however:
- In the generation of action potentials, sodium and potassium ions move through different voltage-gated channels - i.e. the channels are selective for the ion. Transmitter-gated channels usually are not selective for the ions since the pore is too large.
- ion flux through voltage-gated channels is "regenerative" in that increased depolarization caused by the channel opening leads to more influx of the ion. In transmitter-gated channel, the amount of ion passing through the membrane depends on the local concentration of neurotransmitter released at the synapse.
- When acetylcholine causes the opening of the acetylcholine receptor/pore, sodium can flow in and potasium can flow out (down their respective concentration gradients). However, more sodium flows in initially than potassium flows out since potassium efflux is resisted by the negative transmembrane potential. This leads to an inward flow of sodium enough to cause the activation of potential-gated sodium channels. Since these are voltage gated, much more sodium can enter the cell, leading to an action potential and the ultimate opening of vesicular calcium ion channels. This leads to a rise in intracellular calcium (through opening of a Ca channel in membranes of intracellular organelles with high concentrations of Ca ions), which activates muscle contraction.
![]() Electron
Micrograph of Junction-End Plate
Electron
Micrograph of Junction-End Plate
![]() Animation:
Acetylcholine receptor
Animation:
Acetylcholine receptor
![]() Updated
Acetylcholine Receptor Pore
Jmol14 (Java) |
JSMol (HTML5) (1OED)
Updated
Acetylcholine Receptor Pore
Jmol14 (Java) |
JSMol (HTML5) (1OED)
![]() How
does ligand binding lead to channel opening? This has been studied in
detail by Douherty and Lester for binding of acetylcholine, a positively
charged ligand to its neural receptor, the neuronal nicotinic nACh receptor,
a ligand-gated ion channel. It, along with the neural serotonin (HT3)
and GABA receptors belong to the
Cys-Loop superfamily of receptors which consists of 5 protein chains.
Douherty and Lester have reconstituted the AChR into Xenopus oocyte (egg)
membranes and have done a combination of electrophysiological and chemical
studies to probe the conformational changes in the receptor. In the
absence of ligand the channel pore is blocked by 5 leucine side chains,
which move on ligand binding. The neurotransmitter binds to a
tryptophan side chain in an "aromatic box" that contains five aromatic amino
acids), with the positive charge interacting with the aromatic cloud by
cation-π interactions. Serotonin and benzodiazepines (a ligand
that interacts with the GABA channel - see below) bind to different
aromatic side chains in the aromatic boxes in their receptors, indicating
much less specific interactions compared to enzyme-substrate interactions.
This explains how nicotine can bind to neural AChR and activate it even
though their structures differ significantly. They found that the
conformational changes allowing channel opening requires a trans-cis
isomerization of a Pro side chain. Site-specific mutants that favor
the trans form favor closing. However, not all members of the Cys-Loop
superfamily have a Pro at this position.
How
does ligand binding lead to channel opening? This has been studied in
detail by Douherty and Lester for binding of acetylcholine, a positively
charged ligand to its neural receptor, the neuronal nicotinic nACh receptor,
a ligand-gated ion channel. It, along with the neural serotonin (HT3)
and GABA receptors belong to the
Cys-Loop superfamily of receptors which consists of 5 protein chains.
Douherty and Lester have reconstituted the AChR into Xenopus oocyte (egg)
membranes and have done a combination of electrophysiological and chemical
studies to probe the conformational changes in the receptor. In the
absence of ligand the channel pore is blocked by 5 leucine side chains,
which move on ligand binding. The neurotransmitter binds to a
tryptophan side chain in an "aromatic box" that contains five aromatic amino
acids), with the positive charge interacting with the aromatic cloud by
cation-π interactions. Serotonin and benzodiazepines (a ligand
that interacts with the GABA channel - see below) bind to different
aromatic side chains in the aromatic boxes in their receptors, indicating
much less specific interactions compared to enzyme-substrate interactions.
This explains how nicotine can bind to neural AChR and activate it even
though their structures differ significantly. They found that the
conformational changes allowing channel opening requires a trans-cis
isomerization of a Pro side chain. Site-specific mutants that favor
the trans form favor closing. However, not all members of the Cys-Loop
superfamily have a Pro at this position.
In a very recent finding, Dougherthy's group discovered the mechanism underlying the fact that nicotine does not bind with high affinity to the acetylcholine receptor in muscles. Apparently it can not make the cation-π interactions necessary for interaction with the receptor. A H-bond between a H attached to the positive charged nicotine to the Trp carbonyl in the aromatic box allows the cation-π interaction in the neural form. The H bond is not apparently not present in the muscle form, which prevents strong cation-π interaction with nicotine. If it could bind to the muscle acetylcholine receptor, nicotine would be so poisonous that no one would smoke.
B7. Voltage-Gated Sodium Channel
Patch clamp recordings of the sodium channel reconstituted in vesicles shows it to be a voltage-gated channel. Two potent neurotoxins, tetrodotoxin (from Puffer fish) and saxitoxin bind to the channel and act as antagonist (inhibits the activity of the receptor by blocking sodium influx).
Figure: Puffer fish

The guanidino group of the tetrodotoxin appears to bind with high affinity to the entrance of the channel that interacts with the hydrated sodium ion. Affinity chromatography using tetrodoxin beads have been used to purify the protein. The protein sequence consists of 4 repeating units, each containing 6 transmembrane helices. Ion selectively of the sodium channel is determined by the size of the the ion when it is hydrated by one water molecule.
![]() Tetrodotoxin:
Ancient Alkaloid from the Sea
Tetrodotoxin:
Ancient Alkaloid from the Sea
![]() Saxitonin:
From Food Poisoning to Chemical Warfare
Saxitonin:
From Food Poisoning to Chemical Warfare
Figure: Tetrodoxin and Saxitoxin - Structures
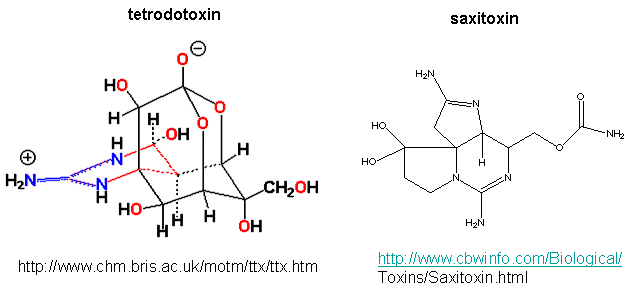
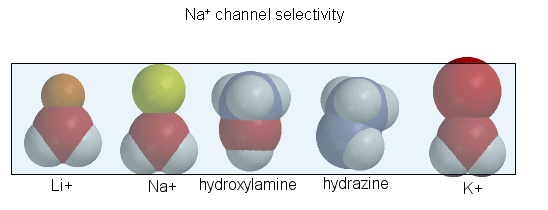
Figure: Ion selectively of the sodium channel
Hence Li and Na ions can pass through, but not the large K ion. Hydroxylamine and hydrazine both have the same width as a water-Na+ complex, and are both positively charged. These can also pass through the channel. At the narrowest part of the channel a water (and presumably the hydroxylamine and hydrazine) form H-bonds to side chains in the pore while the positive charge on the ion forms transient salt-bridges to either a Glu as Asp in the pore. The open channel can likewise spontaneously inactivate by undergoing another conformational change. Comparison of the sequence of this channel with calcium and potassium channels show extensive sequence and structural similarities. One of the 6 transmembrane helices, S4, appears to be the voltage sensor. Depolarization of the membrane potential rmay result in an outward movement and rotation of the positively charged helixes containing Lys and Arg side chains which had presumably formed salt-bridges with negatively charged side chains within the protein. Depolarization of the membrane results in breaking a few salt bridges, and effectively leads to the movement of 1 or 2 charges on the helix through the membrane. Work occurs when charges are moved through an electric field. Work is also related to the ΔG for the system, which is also dependent on the ratio of the open and closed form of the channel. Other voltage-gated ion channels (for potassium and chloride) have a similar membrane topology and an S4 voltage-sensor helix.
B8. Voltage-Gated Potassium Channel
How can the two essential features of the K+
channel pore - selectivity for the larger potassium ion over the sodium ion,
and the near diffusion-limited rate of potassium ion flux through the
channel be reconciled?
Figure: Electrostatics of the K+ channel .
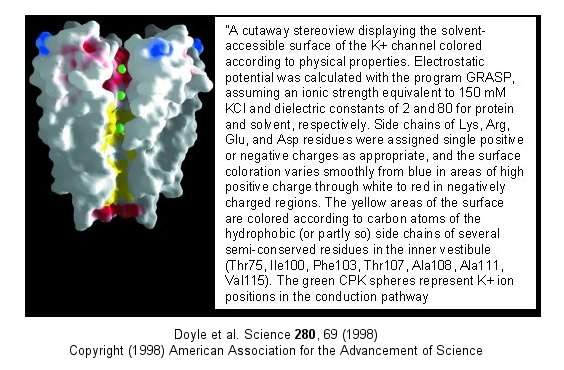
The selectivity filter is composed of many stacked rings of oxygens
that can interact with a dehydrated K ion but not with a dehydrated Na ion
which can not approach close enough to form significant interactions.
Surrounding the filter are twelve aromatic amino acids which constrain the
size of the pore opening. The interactions of the filter O's with the
K ion makes up for the energetically disfavored dehydration of the ion.
The filter contains two K ions which repel each other, assisting in
the vectorial discharge of the ions through the membrane. These ions
must form weak interactions with the selectivity filter. The actual
pore is mostly hydrophobic, which facilitates flow-through of the ions.
The central cavity of the pore can hold some waters molecules in addition to
the K ions which helps stabilize the ion in the middle of the pore.
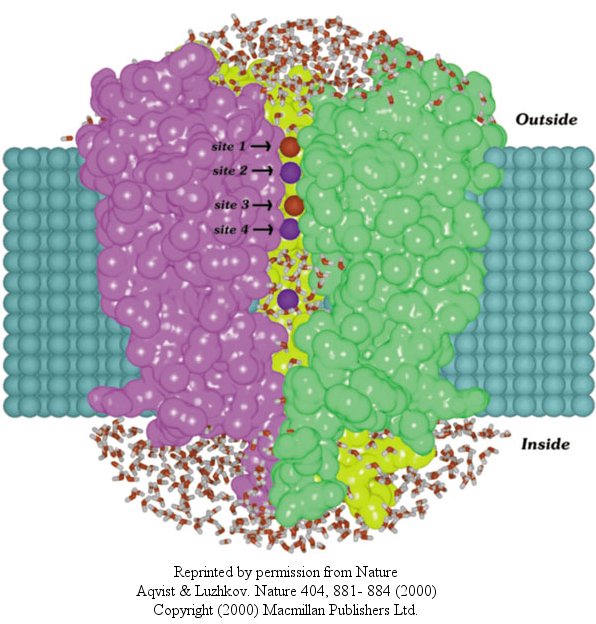
In another recent paper (Nature, 404, April 20, 2000, 881-884), molecular dynamic calculation were performed to distinguish between various models for binding and selectivity of the channel. They found the favored conduction pathway arises from conformational changes between 2 discrete shapes that differ in energy by only about 5 kcal/mol (about 1-2 H bonds). The picture below shows the channel with one of the 4 subunits removed for clarity. Water is shown in red and the K ions in blue. Two ions are in the filter and one in the middle cavity.
Figure: K+ Channel from Steptomyces lividans
![]() Jmol:
Potassium Channel Not updated; Needs work
Jmol:
Potassium Channel Not updated; Needs work
MacKinnon's laboratory (2003) has recently determined the X-ray structure of a voltage-dependent K+ channel from Aeropyrum pernix, an archaebacteria. This protein is homologous to eukaryotic K+ channels. This membrane protein was solubilized with detergent and crystallized in the presence of an antibody which inhibited the flexibility of part of the protein, facilitating the formation of crystals. Like the sodium channel, this protein has 4 subunits each consisting of a 6 alpha-helices which cross the membrane. The S4 helix, which has multiple positively charged side chains, is present in each subunit and acts as a voltage sensor. It was presumed that the helices would would twist in a ratchet-like fashion, leading to the movement of positive charge in the S4 helix across the membrane (as described above for the Na channel). MacKinnon's group found evidence for another mechanism for voltage sensing. Crystal structures show that the S4 helices is imbedded in the membrane, and is not interacting directly with other parts of the protein. It forms an alpha-helical haripin (a paddle shape) which can move through the membrane on change of the transmembrane potential. The topological disposition of paddle (within the membrane) explains the effect of small, lipophilic molecules (like anesthetics nerve toxins, insecticides like DDT) on the channel. His group also labeled the S4 helix with a biotin-17 angstrom linker complex. On change of the transmembrane potential the biotin flips from across the membrane where it can bind to (and be detected by) its natural binding partner, avidin. The authors suggest that the "voltage sensitive paddles operate somewhat like hydrophobic cations attached to levers, enabling the membrane electric field to open and close the pore."
Figure; S4 paddle voltage sensor of the voltage-dependent K+ channel from Aeropyrum pernix

Channels, once open, must be inactivated. In the case of the voltage-gated potassium channel, inactivation occurs when the amino-terminal cytoplasmic domain binds to the potassium pore on the cytoplasmic side, in interaction likened to the binding of a "ball on a chain" (the cytoplasmic domain) to the pore opening. The chain acts to tether the ball domain so it may swing to bind to the pore opening. The ball domain contains both positively charged and hydrophobic regions. Where is the ball domain in the absence of inhibition? Recent studies (Oliver et al.) have shown that a positive domain can bind to proximal phosphatidyinositol 4,5-bisphosphate (PIP2) lipids on the inner leaflet of the membrane bilayer. When so bound, inactivation of the channel is prevented. As you will see in the next section, PIP2 can also be cleaved to form diacylglycerol and inositol 1,4,5-trisphosphate when cells are activated byexternal factors (hormones, growth factors, etc) in the process of signal transduction.
The latest (2005) new structure (Long et al) from MacKinnon's group of the another eukaroytic K+ channel (in its open state) is more consistent with previous expectations. Instead of using antibodies to stabilize the voltage-sensor domains, they crystallized the physiological channel, which in eukaryotes consists not only of 4 identical membrane spanning subunits, but also an addition domain T1 and another protein, beta. The T1 domain consists of the N terminal regions preceding the first membrane-spanning helix, S1, of each of the four monomers of the channel and is located intracellularly at the entrance point of the channel. Four conserved Arg resides on S4, part of the voltage-sensor domains, are hypothesized to move under the influences of forces arising from changes in the membrane's electric field initiated by ion movement through other ion channels in the membrane. Mechanical work is done by the electric field on the voltage sensor as the charged Arg residues are moved through the electric field and the protein conformation is altered. In turn, the S4 and coupled S5 helices of the voltage sensor do mechanical work on the pore by altering its conformation to open/close the pore, specifically at the activation gate of the pore. This seems quite similar to how iron movement into the heme plane in hemoglobin on oxygenation pulls the proximal His on the F8 helix which then transmits a conformation change to other helices in the subunit, leading to cooperative conformational changes in the tightly packed protein. Two of the Arg residue on the S4 sensor are facing the lipid interface while the other two are buried in the voltage-sensor domain. Their work shows that this domain is clearly separate from the pore domain, much as the extracellular ligand-binding domain of neurotransmitter-gated ion channels are separated from the pore domain. In this case, however, the voltage-sensor domain is buried in the membrane.
![]() Updated
Mammalian Voltage-Dependent Shaker Family Potassium Channel
Jmol14 (Java) |
JSMol (HTML5)
Updated
Mammalian Voltage-Dependent Shaker Family Potassium Channel
Jmol14 (Java) |
JSMol (HTML5)
Alterations of potasium channels has recently been implicated in the inhibitory effects of ethanol. Davies, McIntire, et al. studied effects of ethanol on the round worm C. elegans, believing that alcohol-mediated inhibition of neural activity would be conserved across species. Previously it has been shown that the dose-effect curves for behavioral changes is similar for both invertebrates and vertebrates. Ethanol seems to affect many different proteins that would lead to synaptic inhibition, including GABA and glutamate channels and potassium channels. Many different gene products (dopamine D4 receptors, protein kinase C) are associated with increased sensitivity while others (nitric oxide synthase, dopamine D2) are associated with ethanol resistance. These changes suggests the complex pathways are involved in ethanol effects but don't isolate a specific target for its effects.
When C. elegans were exposed to ethanol for brief time periods, ethanol levels rose to values similar to levels seen in intoxicated drivers (0.1%). They isolated mutants that were resistant to ethanol effects (changes in movement and egg-laying behavior). These mutants affected a single gene, slo-1, homologous to the the slowpoke gene in drosophila. The gene in both organisms encoded a BK potassium channel, whose normal function is to repolarize neural membranes. Mutants strains, when transformed with slo-1+ regained ethanol sensitivity. Ethanol appears to directly activate the channel. This would lead to efflux of potassium ions from the worm, hyperpolarizing the neural cells, leading to inhibition of neural activity. Effects in C. elegans were observed at physiologically relevant ethanol, ranging from those that produce euphoria to sedation in humans.
B9. Excitatory Neurotransmitters
Glutamate is a major excitatory neurotransmitter in the brain. Four types of glutamate receptors are found in the central nervous system. They differ in the nature of ligands which bind to the receptor and which act as agonists.
Figure: Glutamate receptor ligands
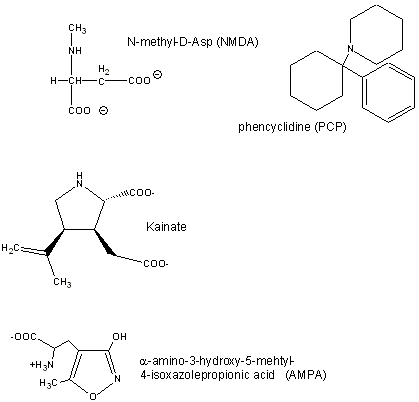
Excessive amounts of glutamate are neurotoxic. Three of the receptor types are briefly described below:
- AMPA receptor binds α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid and acts as a sodium and potassium ion channel.
- Kainate receptor, named after the molecule kainate which acts as an agonist, is a a sodium and potassium ion channel.
- NMDA (N-methyl-D-Asp) receptor allows flow of sodium, potassium and calcium and is regulated by glycine and Zn2+. The street drug PCP (phencyclidine) inhibits this channel.
Sobolevsky et al have recently analyzed the structure, symmetry and mechanism of the rat GluA2 receptor, a homotetramer, a bound competitive antagonist, ZK 200775. GluA2, an AMPA type receptor, has structural similarities to an NMDA type receptor. In order to determine the structure of the wild type GluA2 receptor, a model receptor polypeptide that was termed GluA2cryst (PDB# 3KG2) was crystallized. Isolated GluA2 subunits were used to piece together a detailed model of rat GluA2 as it would exist in its natural membrane bound form. The receptor has three main domains that are essential for its function. The first is a large extracellular amino-terminal domain (ATD). This domain serves as a guide for ligand binding in the next domain, the ligand-binding domain (LBD). The ligand binding domain participates in agonist/antagonist competitive binding that regulates the gating of the ion channel. The final domain is the transmembrane domain (TMD) that forms the ion channel for molecules to pass through the cell membrane.
The GluA2 receptor is shaped like a “Y” with the domains layered on top of each other. The TMDs form the transmembrane section and base on the outside of the membrane. The ATDs protrude outward like the “arms” of a Y and the LBD is on top of the TMD. This process gates the transmembrane channel as can be seen in Fig. 1. Analysis of the crystallized protein uncovered a bound antagonistic ligand proving that the ligand binding occurs in a domain (LBD) rather than between domains, as was previously conceived.
The authors studied the gating mechanism between the LBDs and TMDs by analyzing the change in symmetry that occurs when the gate is opened and closed. When the gate is closed, the LBDs exhibit a two-fold symmetry and the TMDs four-fold symmetry. When the ligand binds to the LBDs an abrupt structural change occurs creating four fold symmetry in the LBDs. This transition was measured by calculating the r.m.s.d. of the alpha carbons between the subunits undergoing the structural change. The TMDs also underwent a conformational change when ligand binding occured and ions travelled down the channel. In order to accommodate the ion transfer, several subunits within the TMDs loosened their helix and provided an open pathway for the ion to travel in to the cytoplasm of the cell.
B10. Inhibitory Neurotransmitters:
The main inhibitory neurotransmitters are GABA (gamma-aminobutyric acid, which is made from Glu through decarboxylation of the α-C group) and glycine. The bind to transmiitter-gated chloride channels, which when open hyperpolarize the membrane. Benzodiazepines (like Valium and Librium - anti-anxiety and muscle-relaxing agents) and barbituates (like phenobarbital-hypnotics) bind at allosteric sites and potentiate the binding of each other and GABA.
![]() Updated
Chloride Channel
Jmol14 (Java) |
JSMol (HTML5)
Updated
Chloride Channel
Jmol14 (Java) |
JSMol (HTML5)
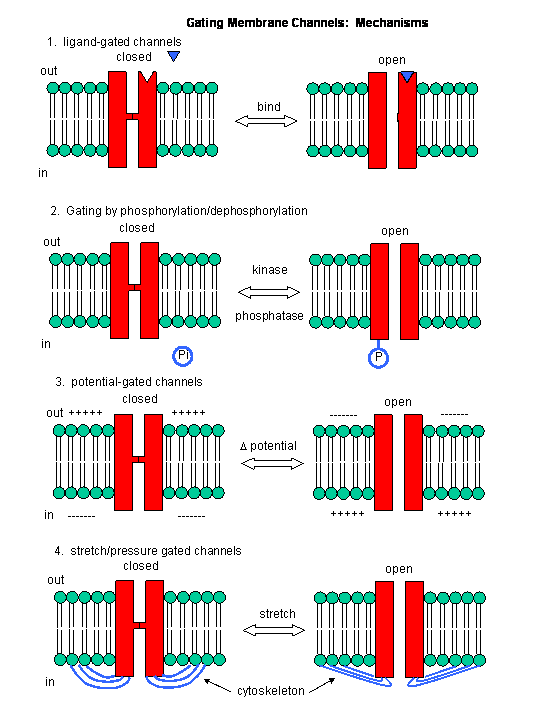
Figure: Mechanisms of Channel Gating
B11. The Senses
Taste (Gustatory)- There are 5 different types of tastants perceptible to mammals. These include sweet, sour, salty, bitter, and umami (taste of monosodium glutamate). It appears that different cells expressing different tastant receptors (TR) mediate taste. These receptors could be ligand-gated ion channels, or work through ligand-induced conformational changes in the receptor which are sensed by intracellular proteins (usually G proteins, that are discussed in the next chapter section).
Sweet - Cells that recognize and respond to sweet tastants express receptors from the TIR family. The receptor appears to be active as a heterodimer. Dimers of T1R2 and T1R3 recognize sucrose and saccharin. The bound receptor is coupled to intracellular G proteins.
Sour - This sense would protect us from excessively acidic foods. Huang et recently found an ion channel, PKD1L1 (Polycystic-Kidney-Disease-Like Ion Channel), that is necessary in mice for the sour sensation. Mice breed to lack this gene could not detect sour stimuli.
Salty - Taste cells with receptors necessary for salt sensation have not yet been identified.
Bitter - Bitter taste perception, required to prevent the ingestion of toxic substances, is mediated by the T2Rs family of receptors, mediated by the G-protein, gustducin.
Umami - The G-protein coupled receptor for this taste consists of a dimer of TIR1 and T1R3.
Smell (Olfactory) - tba
Sight (Visual)- TBA
B12. Touch
A theme of this course has been that binding interaction initiate biological activity. These include substrate binding to enzymes, transcription factors binding to other proteins and DNA, and signaling molecules binding to membrane receptors. These interactions allow free or membrane-bound macromolecules to "sense" their microenvironment. Odorants and tastants also initiate biological responses by binding to membrane receptors (usually serpentine receptors coupled to G proteins). Sight, another sense, is initiated by "binding" (of course I mean interaction with) a photon. But what about touch? Kung has recently written a review to discuss mechanisms involved in mechanosensation, which is ubiquitous in the biological world. He gives many examples in which a biological response is initiated by "touch", including these more uncommon examples:
- worm and paramecium movement,
- plant root response to gravity (gravitropism)
- ratio of height to girth growth in stems to wind and rain (thigmomorphogenesis),
- blood pressure changes (animal baroreceptors),
- muscle stretching (spindle receptors),
- limb position (proprioreceptors)
- texture (receptors in tongue),
- hearing, and
- osmotic pressure changes.
First we will consider the more studied mechanosensitive (MS) ion channels, which respond to changes in turgor induced by osmotic changes in bacterial cells. When reconstituted in purely lipid bilayer vesicles, these proteins can detect forces applied to the membrane. Analogous receptors have been found in animals cells as well. These channels appear to detect changes at the interface between the protein and lipid, and are probably lipid-gated channels.
Consider changes in a bacterial cell placed in pure water. Influx of water into the cells produces a turgor in the cell that causes a great force on the membrane. Cells response by releasing small ions and molecules from the cell (95% of free Pro, for example). Membrane protein involved in this process are called mechanosensitive channels of large and small conductance (MscL and MscS). These channel proteins are nonselective (compared to usual ion channels which demonstrate ion specificity). They retain activity (force induced permeability) in liposomes even if only one type of lipid constitutes the membrane.
Since the lipid bilayer, compared to the bulk solution, is highly ordered (or anisotropic) in an directional sense, it displays a gradient of physical properties at different depths into the membrane. As water displays a significant surface tension (since hydrogen bond-induced forces on water differ for bulk interior water and surface water), so does the bilayer have a significant surface tension between the polar head groups and nonpolar tails. These forces are counterbalanced by other to produce a thermodynamically stable bilayer. Membrane proteins imbedded in the bilayer would experience these forces, and change in interfacial lipids around the protein would be expected to alter the forces on the protein and provide a mechanism for lipid gating of the channels. Steered molecular dynamic simulations (in the sense that forces can be applied at specific locations in a protein) of the MscL imbedded in a bilayer have shown that force channels applied on the protein at the headgroup/acyl chain interface caused a transient opening in the channel. Translational or flip-flop diffusion of lipids to the channel/lipid interface, or addition to the membrane of hydrophobic molecules, such as anesthetics, or amphiphilic molecules, such as phosphatidyl choline with one fatty acid remove or arachidonic acid (both of which can arise in signal transduction processes) can induce opening as well. Gating of the channel appears to require changes in lipid composition in just one leaflet, which would induce localized shape changes such as bulges or cavitations that would alter membrane forces.
Similar proteins have been found in plants and animals. The mammalian TREK-1 potassium channels can be opened by trinitrophenol (an amphiphathic bulge former or crenater) and closed by chloropromazine (an amphipathic cavity-former or cavitator). Single chain amphiphiles like lysophosphatidic acid, which can be modeled like a cone, is an activator (by crenation). Many anesthetics activate it as well. Although structural cytoplasmic protein scaffolding (the cytoskeleton) may be associated with these membrane channels, their involvment in force change in the channel protein and concomitant opening/closing of the channel may not be necessary. Another set of similar proteins, transient receptor potential (TRP) proteins, which sense vibration and other stimuli, have been recently studied. There appears to be many subfamilies of these proteins. Sensory hair cells, responsible for sound transduction, appear to transmit force to a TRP protein through a tether, presumably protein constituents of the cytoskeleton.
Kung argues that two types of cell membrane sensors were required in early cells, those that recognize changes in solute concentration through a "lock and key" mechanism, and those that recognize changes in solvent concentration, which would occur under conditions of excess (rain) and depletion (drought). These changes would be transmitted to membrane sensors not by a lock and key binding mechanism (given the small change in water concentrations from the pure state of 55.5 M, but by osmotic pressure changes affecting the turgor of the cell.
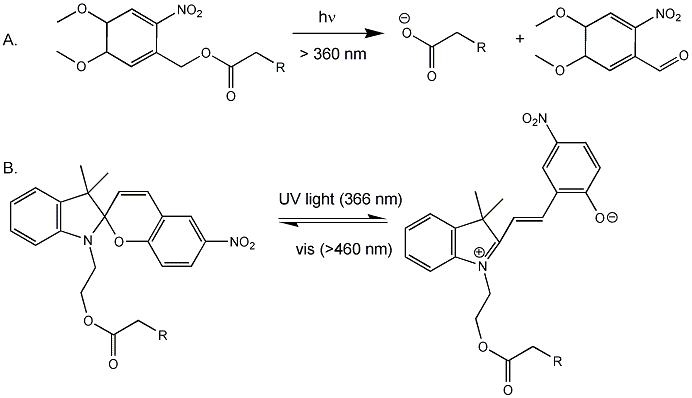
Back to the bacterial MscL for a moment. It consists of 5 identical subunits, each with 2 transmembrane alpha helices, M1 and M2. The pore is lined with the M1 subunits from each subunit. Mutagenesis or covalent modification of glycine 22 in the M1 helix to a polar or charged substitutent causes increase in pore size and hydration in the pore. Kocer et al changed the Gly to a Cys (no other Cys are found in the protein). Then they modified the Cys with an photosensitive ester analog of iodoacetic acid (Figure A below, where R = I). When illuminated with long UV (366 nm), the ester cleaves (photolysis) to form a negatively charged carboxylate and a product with a polar end near the carboxylate. This induced charge separation mimics conformational changes induced in the protein during osmotic shock, causing an opening in the pore when reconstituted into artificial bilayers. The photolysis is irreversible. They then tried another modification (Figure B below) in which the attached molecule is not photolyzed into two separate molecules, but reacts through ring opening to a form with separate + and - charges. This species could be irradiated with visible light and return to the original structure. They effectively made a "light-actuated" nanogate or nanovalue in the channel which allowed flux of ions across the membrane. To test the usefulness of this nanoengineering feat, they made liposomes with a fluorophore inside and the light-gated modified MscL protein in the membrane. The encapsulated fluorophore in the liposome self-quenches when present at high concentration. Upon irradiation, the pore opened as reflected by increased fluorescence when fluorophore levels inside the liposome decreased, which reducing fluorescence quenching.
Figure: Light-Activated Nanovalve in MscL
![]() Movie:
Steered MD simulation of MscL
Movie:
Steered MD simulation of MscL
B13. Summary
1. the same ions moving through different channels can have different consequences. Consider the example of potassium ions. When it moves through a:
- nongated channel, it establishes the resting potential
- a voltage gated channel, it repolarizes the cell after an action potential
- channel that is gated by chemical modification (such as phosphorylation), it may hyperpolarize the membrane (make it more negative inside) and facilitate neuron inhibition
2. If two ions are involved in signaling, the result depends on
- if the two species move simultaneously through the same channel (which results in excitation through an initial depolarization of membrane if the ions are potassium and sodium)
- if the two species move through different channels in a sequential fashion (which generates an action potential) if the ions are potassium and sodium).
In summary, ligand and voltage gated channels allow changes in the polarization of the membrane. Other mechanisms can also lead to changes. Membrane proteins can be phosphorylated (using ATP) by protein kinases in the cell, leading to a change in the conformation of the membrane protein, and either an opening or closing of the channel. Channels linked to the cytoskeleton of the cells can also be opened or closed through stretching. Other stimuli that gate channels are light (through photoisomerization-induced conformational changes), heat, and cold.
B14. Links and References
![]() MetaNeuron:
a dowloadable program to simulate neural activity (from the U.MN)
MetaNeuron:
a dowloadable program to simulate neural activity (from the U.MN)
![]() Ligand-Gated
Ion Channel Database
Ligand-Gated
Ion Channel Database
![]() Senomyx:
Flavors and Taste Receptors
Senomyx:
Flavors and Taste Receptors
![]() Animation:
Comparison of direct and indirect neurotransmitter actions, ligand-gated
vs. G-protein-coupled receptor.
Animation:
Comparison of direct and indirect neurotransmitter actions, ligand-gated
vs. G-protein-coupled receptor.
References
-
Sobolevsky, A. et al. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 729, 745 (2009)
-
Xie, Xinan et al. Nicotine binding to brain receptors requires a strong cation-π interaction. Nature doi:10.1038/nature07768 (published March 09)
-
Sarah C. R. Lummis, Darren L. Beene, Lori W. Lee, Henry A. Lester, R. William Broadhurst, Dennis A. Dougherty. Cis/trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. 438, 248 - 252 (2005),
-
Everts, S. Neuron Activation. C&E News. 44, Oct 2, 2006
-
Huang, A. et al. The cells and logic for mammalian sour taste detection. Nature 442, pg 934 (2006)
-
Long, S. B. et al. Crystal Structure of a mammalian voltage-dependent Shaker family K+ Channel. Science 309, 897 (2005); Long, S. B. et al. Voltage Sensor of KV1.2: Structural Basis of Electromechanical Coupling.
-
Kung, C. A possible unifying principle for mechanosensation. Nature 436, 647 (2005)
- Oliver, D. et al. Functional Conversion Between A-Type and Delayed Rectifier K+ Channels by Membrane Lipids. Science. 304, pg 265 (2004)
- Davies, A. et al. A Central Role of the BK Potassium Channel in Behavioral Response to Ethanol in C. Elegans. Cell, 115, pg 655 (2003)
- Miyazawa, A. et al. Structure and Gating mechanism of the acetylcholine receptor pore. Nature, 423, pg 949 (2003)
- Jiang, Y, MacKinnon, R. et al. X-ray structure of a voltage-dependent K+ channel. Nature. 423, pg 33 (2003); The Principle of gating charge movement in a voltage-dependent K+ channel. Nature. 423, pg 42 (2003)
- Kandel et al. Principles of Neural Science, 3rd edition. Elsevier (1991). This is the source for much of the material in this section.
- Schumacher and Adelman. An Open and Shut Case. (summairzing work on Ca2+-gated K channel from MacKinnon's lab) Nature. 417, pg 501 (2002)
- Sun et al. Mechanism of glutamate receptor desenitiziation. Nature. 238. pg 238, 245 (2002)
- Dutzler et. al. X-ray structure of a CIC Chloride Channel at 3.0 Angstroms Reveals the molecular basis of anion selectivity. Nature. 415, pg 276, 287 (2002)
- Berneche et al. Energetics of Ion Conductions Through the K+ Channel. (use of dynamics) Nature. 414 p 23, 73 (2001)
- Sixma et al.. Snails, synapses, and smokers (nicotine/receptor). Nautre, 411, pg 252 (2001)
- MacKinnon et al. 50 Years of Inactivation (How K+ channels are closed). Nature. 411, pg 643, 657 (2001)
- McGaugh, J. Memory - a century of consolidation. Science. 287. pg 248 (2000)
- Wilson et al. Cannabinoids act backward. (drugs that control signaling from cannabinoids) Nature. 410, pg 527, 588 (2001)
- McGaugh. Memory - A century of consolidation. Science. pg 248 (2000)
- Olney et al. How alcohol damages the brain (Fetal alcohol syndrome). Science. 287, pg 947, 1056 (2000)
- Hardy et al. Genetic Classification of Primary Neurodegenerative Disease. Science 282, pg 1075 (1998)
- Greenberg et al. Learning more about the NMDA receptor regulation (involved in learning and memory). Science. 295. pg 449, 491 (2002)
- Meshorer et el. Alternative Splicing and neuritic mRNA translocation under long-term neuronal hypersensitivity (in PTSD) Science. 295. pg 508 (2002)
Navigation
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 9B: Neural Signaling

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.