Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 6 - TRANSPORT AND KINETICS
A: PASSIVE AND FACILITATED DIFFUSION
BIOCHEMISTRY - DR. JAKUBOWSKI
Last Update: 04/08/16
|
Learning Goals/Objectives for Chapter 6A: After class and this reading, students will be able to
|
A2. Facilitated Diffusion
Consider the following mechanism:
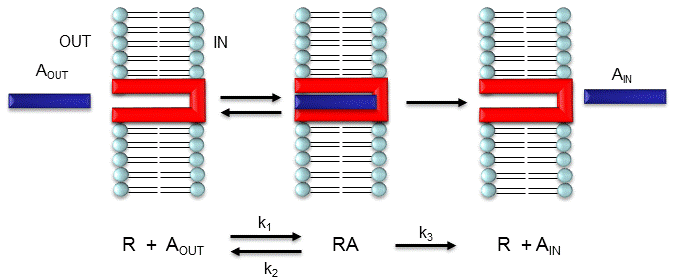
Let's assume that for this system the initial flux will be measured. We would like to derive equations which show J as a function of Aout (assuming that Ain is negligible over the time course of measuring the initial flux. Also assume that the J facilitated is much greater than J passive. In contrast to passive diffusion, JA is not proportional to Aout but rather to [Abound].
Consider this example to help you understand that proportionality. Pretend that the receptor is a truck which can carry one particle across the membrane at a time (i.e. 1/1 stoichiometry). Also assume that the particle can't get across without being carried by the truck. If there are no trucks in the membrane, no load can be delivered. If there are trucks in the membrane but no particles in them, no load will be delivered. As the number of particles available to be loaded into the truck increase, the truck will have an increased chance to be loaded (depending of course on the affinity of the particle for the truck). If the number of loaded trucks is doubled, the number of particles dumped to the other side will double. Therefore, by analogy,
JA is proportional to [RA], or
Equation 1: JA = const [RA] = k3 [RA]
How can we calculate RA when we know A and R? Let us assume that Atotal (A0) is much greater than R0, as is the likely biological case, and Ain = 0. We can calculate RA using the following equations, and the same procedure we used for the derivation of the binding equation
ML = (MoL)/(Kd + L)
Equation 2 - Dissociation constant: K d = ([A]eq[R]eq)/[AR]eq = ([A][R])/[RA] (units of molarity)
Equation 3 - Mass Balance of R: Ro = R + RA so R = Ro -RA
Since we will assume that A0 is much greater than R0, we will not need the mass balance for A (which is Ao = A + RA)
Substitute 3 into 2:
(RA)Kd = (Ro)A - (RRA)A
(RA)Kd + (RA)A = (Ro)A
Equation 4: RA = (Ro)A/(Kd + A)
Substitute 4 into 1 gives the final equation,
Equation 5: JA = const [RA] = k3 [RA] = k3 (Ro)A/(Kd + A).
It should be clear to you from this equations that:
- a plot of JA vs A is hyperbolic
- JA = 0 when A = 0.
- JA = Jmax when A is much greater than Kd
- A = Kd when JA = Jmax/2.
![]() Wolfram
Mathematica CDF Player - Jo vs A (free
plugin required)
Wolfram
Mathematica CDF Player - Jo vs A (free
plugin required)
![]() Interactive SageMath
Graph: ML vs L at different Mo and Kd values
Interactive SageMath
Graph: ML vs L at different Mo and Kd values
These are the same conditions we detailed for our understanding of the binding equation
ML = (MoL)/(Kd + L)
RAPID EQUILIBRIUM ASSUMPTION
This derivation is based on the assumption that the relative concentrations of A, R, and AR can be determined by the Kd for the interactions and the concentrations of each species during the early part of diffusion (i.e. under initial rate conditions). Remember under these conditions, Aout does not change much with time. Is this a valid assumption? Examine the mechanism shown in the above figure. Aout binds to R with a second order rate constant k1. RA has two fates. It can dissociate with a first order rate constant k2 to Aout + R (to give the original species), or dissociate with a first order rate constant of k3 to give Ain + R (as A moves across the membrane). If we assume that k2 >> k3 (i.e. that the complex falls apart much more quickly than A is carried in), then the relative ratios of A, R, and RA can be described by Kd. Alternatively, you can think about it this way. If A binds to R, most of A will dissociate, and a small amount will be carried across the membrane. If this happened, then R is now free, and will quickly bind Aout and reequilibrate. This occurs since the most likely fate of bound A is to dissociate, not to be carried across the membrane, since k3 << k2.
Diffusion of Ions and Molecules Across Membranes
As we saw previously, the permeability coefficients of synthetic membranes (liposomes) to solutes is related to the size and polarity of the solute.
Figure: permeability coefficients
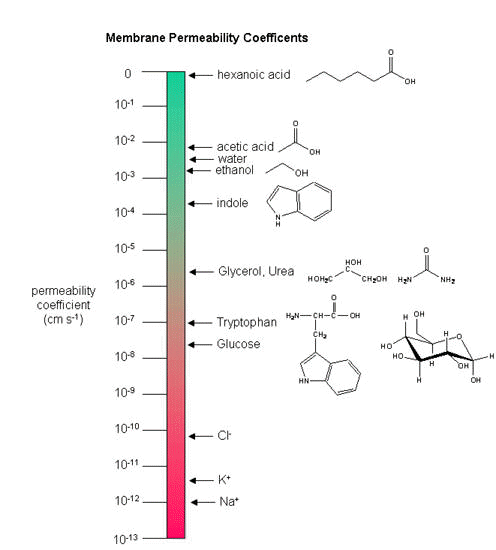
Smaller, higher charge density ions (like Na+) have a lower permeability coefficient than do larger, lower charge density ions (like K+). What about natural membranes? Look at the chart below.
Permeability coefficient (cm/s) of natural and synthetic membranes to D-Glucose and D-mannitol at 25oC.
| Membrane Preparation | D-Glucose | D-Mannitol |
| Synthetic Lipid Bilayer | 2.4 x 10-10 | 4.4 x 10-11 |
| Calculated Passive Diffusion | 4 x 10-9 | 3 x 10-9 |
| Intact Human Erythrocyte (red blood cell) | 2.0 x 10-4 | 5 x 10-9 |
Why do the permeability coefficients differ for D-Glc and D-Mannitol across the red blood cell membrane?
Navigation
Return to Chapter 6A: Passive and Facilitated Diffusion Contents
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 6A: Passive and Facilitated Diffusion

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.