Cut Site Selection by the Two Nuclease Domains of the Cas9 RNA-guided Endonuclease. Hongfan Chen, Jihoon Choi and Scott Bailey.The Journal of Biological Chemistry, 289, 13284-13294 (2014)
3. The investigators then studied their system in vitro by reconstituting the components. They monitored DNA cleavage by LMG18311 Cas9 as a measure of activity.
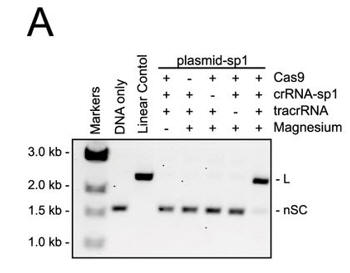
a. Figure 3A shows an electrophoresis gel of reaction mixtures containing 5 nm target plasmid, 25 nm Cas9, 25 nm crRNA (42-nucleotide crRNA containing the sequence derived from first spacer of CRISPR-1), 25 nm (42-nucleotide) tracrRNA, and 10 mm Mg2+ that were incubated for 30 min at 37 °C. The agarose gels were stained with ethidium bromide. Interpret the results. Why does the linear control migrate less far than the uncut plasmid?

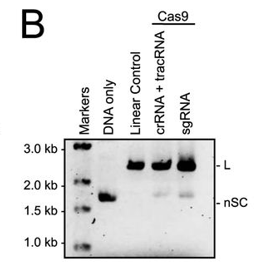
b. In Figure B, a cognate sgRNA was substituted for separate crRNA and tracrRNA molecules. Interpret the gel results..
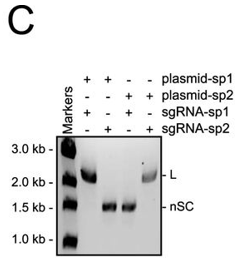
c. In Figure C, they studied cleavage in the presence of different sgRNA sequences. Explain the results.
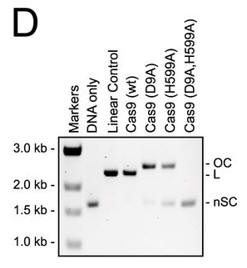
d. For figure D, the used different active site mutants of Cas 9. The (D9A) mutant was in the RuvC-like domain while the H599A mutant was in the HNH domain Explain the results. What effect does the double mutant have on the activity?
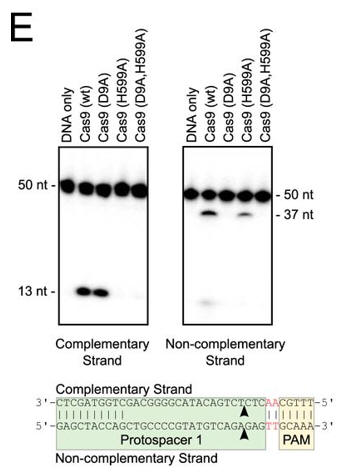
e. In Figure E, the authors studied which domain was involved in the cleavage of each strand (complementary to the guide DNA or the noncomplementary strand) of the target DNA. The dsDNA was radiolabeled at the 5′ end of the complementary strand (left hand gel) or the noncomplementary strand (right hand gel). Reactions were performed as in A, and products were separated by 10% denaturing PAGE. The cleavage sites are indicated with arrows in the schematic diagram (bottom). 50 nt, 50 nucleotides; 37 nt, 37 nucleotides. Interpret the results.
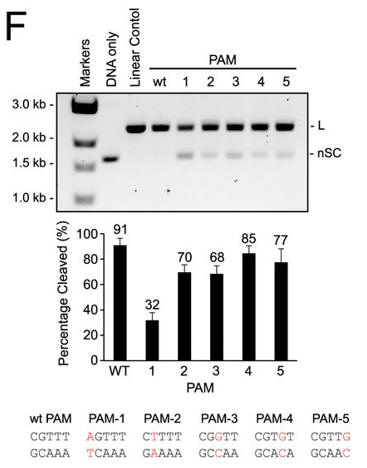
f. Do mutations in the PAM or changes in linker length have the same effect on DNA interference in vitro as they did in vivo? The investigators monitored cleavage of these variant plasmids by recombinant LMG18311 Cas9. Interpret the results in Figure F.
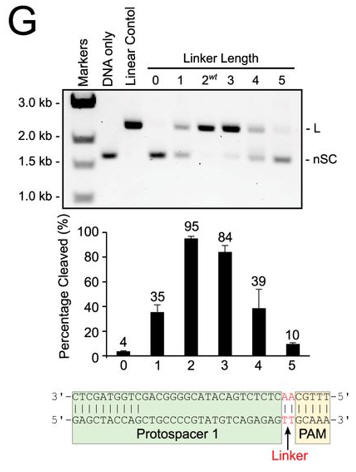
g. In Figure G, cleavage of plasmid targets containing the indicated linker lengths was studied. In A–C and E–F, the positions of negatively supercoiled (nSC), linear (L) and nicked or open circle (OC) plasmid are indicated. The linear control is a digestion of the plasmid target with the restriction enzyme AgeI
h. In the mutational analysis, they substituted asparatic acid for alanine. Draw the structures of the side chains of D and A. Why did they choose A to substitute for D? What role might D have in catalysis in the wild-type protein?