Cut Site Selection by the Two Nuclease Domains of the Cas9 RNA-guided Endonuclease. Hongfan Chen, Jihoon Choi and Scott Bailey.The Journal of Biological Chemistry, 289, 13284-13294 (2014)
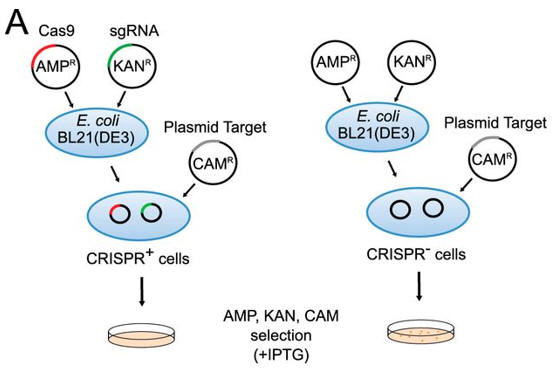
2. To confirm that the identified PAM was functional, investigators needed a selection system. They used a transformation assay in which E. coli cells containing an exogenous type II CRISPR-Cas system were resistant to plasmid transformation, whereas cells lacking the system are competent for transformation. Explain how this system could be used to determine if specific PAM are functional?
ANSWER: The E. Coli cells with an added CRISRP-Cas system should be able to cleave any plasmid DNA taken up by the cell as the plasmid DNA would serve as the "target" much like phage DNA. Cells without the exogenous CRISRP-Cas system could not cleave plasmid that were taken up by the cell, which could allow the cells with intact plasmid to be transformed by specific genes brought in and expressed by the plasmid.
a. Figure A shows a schematic representation of transformation assay.

The target plasmid contained protospacer-1 (whose sequence was identical to the first spacer of CRISPR-1), a 2-bp linker, and the identified PAM. The first control plasmid contained only protospacer-1, whereas the second control plasmid lacked both protospacer-1 and PAM. The target and control plasmids were then used to transformed cells. in the presence of IPTG and the appropriate antibiotics.
Why did the investigators add IPTG? What difference would you expect to find in the cells in the left panel compared to the right in their assay?
Answer: Protein expression of the Cas9 protein is necessary for a function CRISPR system. Presumably, the protein is under the control of a lac promoter which is turned out with lactose, allolactose, or IPTG. The cells on the left, which have a functional Cas9 protein and sgRNA will be able to cleave the plasmid that would provoke cell transformation and hence they would not be transformed.
b. Figure 2B shows plasmid transformation by LMG18311 Cas9 and sgRNA in E. coli cells. Transformation efficiency is expressed as cfu per 5 ng of plasmid DNA. Average values from at least three biological replicates are shown, with error bars representing 1 S.D. Interpret and explain results.
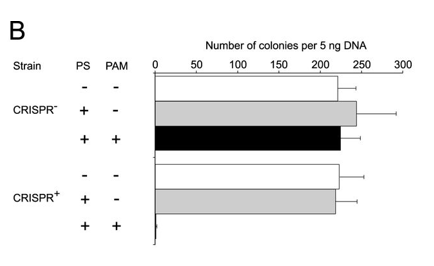
ANSWER: The control plasmids transformed into both strains with similar efficiency (Fig. 2B). The target plasmid failed to transform into the CRISPR+ cells but transformed into the CRISPR−. this shows that a function PAM seqeunce is required for the Cas9 protein to cleave the plasmid and block transformation.
c. For Figure C, the investigators made single nucleotide changes in the PAM sequence of the CRISPR+ plasmid and studied the effect on plasmid transformation efficiency. Interpret he results.
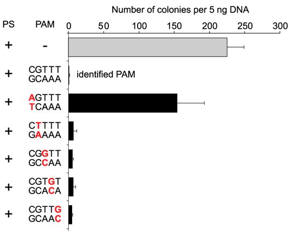
Answer: . Only the plasmid containing a mutation at the position 1 guanosine (that is, the PAM nucleotide closest to the protospacer) was transformed, albeit with a reduced (abouy 66%) transformation efficiency as compared with the intact PAM sequence . Plasmids containing single mutations to any of the other four positions were resistant to transformation. These results indicate that the guanosine at position 1 is important for PAM function but individually the four other positions have little effect on PAM function.
d. For Figure D, the effect of the length of the linker length on plasmid transformation efficiency was studied. PS denotes the protospacer sequence. Interpret the results.
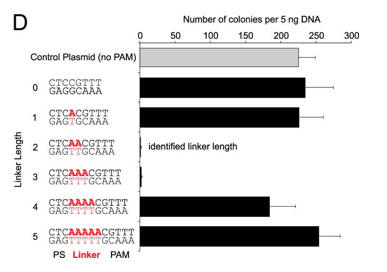
The CRISPR+ cells were equally resistant to transformation by a plasmid target with a linker length of either 2 bp or 3 bp. Plasmids with other linker lengths transformed with efficiencies more similar to the control plasmid, suggesting that plasmids with these linkers were able to escape CRISPR-Cas silencing.