Structure & Reactivity in Chemistry
Introduction to Biomolecules
IB3. Intermolecular Forces in Biology: Nucleic Acids (contributed by Henry Jakubowski)
The nucleic acids RNA and DNA are involved in the storage and expression of genetic information in a cell. Both are polymers of monomeric nucleotides. DNA exists in the cell as double-stranded helices while RNA typically is a single-stranded molecule which can fold in 3D space to form complex secondary (double-stranded helices) and tertiary structures in a fashion similar to proteins. The complex 3D structures formed by RNA allow it to perform functions other than simple genetic information storage, such as catalysis. Hence most scientists believe that RNA preceded both DNA and proteins in evolution as it can both store genetic information and catalyze chemical reactions.
DNA
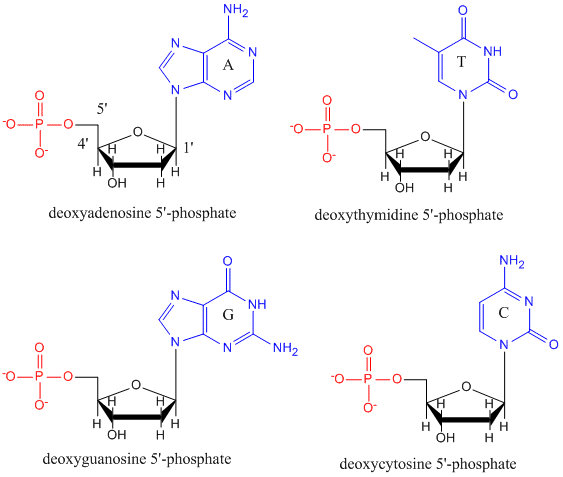
DNA is a polymer, consisting of mono
mers call deoxynucleotides. The monomer contains a simple sugar (deoxyribose, shown in black below), a phosphate group (in red), and a cyclic organic R group (in blue) that is analogous to the side chain of an amino acid.

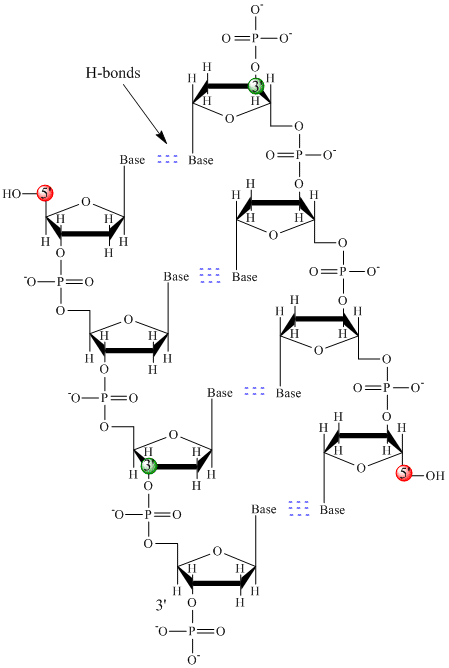
Only four bases are used in DNA (in contrast to the 20 different side chains in proteins) which we will abbreviate, for simplicity, as A, G, C and T. They are bases since they contain amine groups that can accept protons. The polymer consists of a sugar - phosphate - sugar - phosphate backbone, with one base attached to each sugar molecule. As with proteins, the DNA backbone is polar but also charged. It is a polyanion. The bases, analogous to the side chains of amino acids, are predominately polar. Given the charged nature of the backbone, you might expect that DNA does not fold to a compact globular (spherical) shape, even if positively charged cations like Mg bind to and stabilize the charge on the polymer. Instead, DNA exists usually as a double-stranded (ds) structure with the sugar-phosphate backbones of the two different strands running in opposite directions (5'-3' and the other 3'-5'). The strands are held together by hydrogen bonds between bases on complementary strands. Hence like proteins, DNA has secondary structure but in this case, the hydrogen bonds are not within the backbone but between the "side chain" bases on opposing strands. It is actually a misnomer to call dsDNA a molecule, since it really consists of two different, complementary strands held together by hydrogen bonds. A structure of ds-DNA showing the opposite polarity of the strands is shown below.
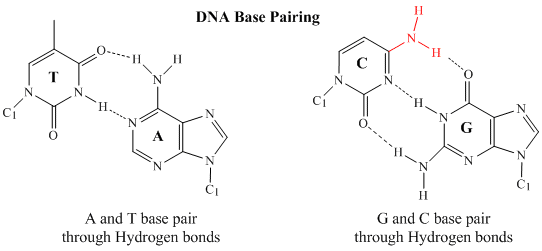
In double stranded DNA, the guanine (G) base on one strand can form three H-bonds with a cytosine (C) base on another strand (this is called a GC base pair). The thymine (T) base on one strand can form two H-bonds with an adenine (A) base on the other strand (this is called an AT base pair). Double-stranded DNA has a regular geometric structure with a fixed distance between the two backbones. This requires the bases pairs to consists of one base with a two-ring (bicyclic) structure (these bases are called purines) and one with a single ring structure (these bases are called pyrimidines). Hence a G and A or a T and C are not possible base pair partners.
Double stranded DNA varies in length (number of sugar-phosphate units connected), base composition (how many of each set of bases) and sequence (the order of the bases in the backbone). The following links provide interactive Jmol models of dsDNA made by Angel Herráez, Univ. de Alcalá (Spain) and Eric Martz.
![]() External JSmol:
External JSmol:
Chromosomes consist of one dsDNA with many different bound proteins. The human genome has about 3 billion base pairs of DNA. Therefore, on average, each single chromosome of a pair has about 150 million base pairs and lots of proteins bound to it. dsDNA is a highly charged molecule, and can be viewed, to a first approximation, as a long rod-like molecule with a large negative charge. It is a polyanion. This very large molecule must somehow be packed into a small nucleus of a tiny cell. In complex (eukaryotic) cells, this packing problem is solved by coiling DNA around a core complex of four different pairs (eight proteins total) of histone proteins (H2A, H2B, H3, and H4) which have net positive charges.
The histone core complex with dsDNA wound around approximately 2.5 times is called the nucleosome.
![]() Jmol: Nucleosome
Jmol14 (Java) |
JSMol (HTML5)
Jmol: Nucleosome
Jmol14 (Java) |
JSMol (HTML5)
DNA can adopt two other types of double-helical forms. The one discovered by Watson and Crick and found in most textbooks is called B-DNA. Depending on the actual DNA sequence and the hydration state of the DNA, it can be coaxed to form two other types of double-stranded helices, Z and A DNA. The A form is much more open then the B form.
The 3.2 billion base pairs of DNA in humans contains about 24,000 short stretches (genes) that encode different proteins. These genes are interspersed among DNA that helps determining if the gene is decoded into RNA molecules (see below) and ultimately into proteins. For a particular gene to be activated (or "turned on"), specific proteins must bind to the region of a particular gene. How can binding proteins find specific binding targets among the vast number of base pairs that to a first approximation have a repetitive sugar-phosphate-base repeat? The Jmol below shows how specificity can be achieved. When DNA winds into a double helix through base-pairs between AT and GC, hydrogen bond donors (amide Hs) and acceptors (Os) on the bases that are not used in intrastrand base pairing,are still available in the major and minor grove of the ds-DNA helix (see Jmol below). Unique base pair sequences will display unique patterns of H bond donors and acceptors in the major grove. These donors/acceptors can be recognized by specific DNA binding proteins which on binding can lead to gene activation.
![]() Jmol:
dsDNA showing unique H bond donors and acceptors in the
major grove
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
dsDNA showing unique H bond donors and acceptors in the
major grove
Jmol14 (Java) |
JSMol (HTML5)
RNA
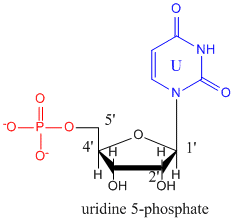
The monomers that make up RNA are identical to those for DNA with two differences. The 2'C of the ribose ring has an OH instead of H as is found in deoxyribose. In addition, the thymine base found in DNA is replaced with uracil. The structure of uridine-5-monophosphate is shown below.
Based on analogy with DNA, you might expect RNA to adopt a double-stranded helical conformation. It can do this in short stretches, but instead, even in the presence of positively charged divalent cations, it is found predominantly in a single stranded form that folds to form regions of secondary structure (double stranded helices) imbedded in a complex tertiary structures (not unlike a protein).
The addition of the 2'OH on the ribose ring or RNA alters the pucker of the ribose ring and through steric interference destabilizes ds-RNA in comparison to ss-RNA. The five-membered ribose or deoxyribose ring can adopt an envelope-like structure in which one ring atom, representing the tip of the envelope, is above, or endo, to the other four coplanar atoms. In deoxyribose, the 2' ring carbon atom prefers the endo position while in ribose, the 3' C is endo (see Jmol model below). When double-stranded RNA forms, it is actually very similar in structure to the more open and less abundantly found A-DNA form.
![]() Jmol:
Jmol model showing puckering of ribose ring of RNA and deoxyribose ring of
DNA (updated to HTML5 asap)
Jmol:
Jmol model showing puckering of ribose ring of RNA and deoxyribose ring of
DNA (updated to HTML5 asap)
![]() Jmol:
Jmol model comparing dsDNA and dsRNA
(updated to HTML5 asap)
Jmol:
Jmol model comparing dsDNA and dsRNA
(updated to HTML5 asap)
Here are some examples of complex tertiary structures formed from single stranded RNA which folds to form a compact 3D structure with some double-stranded A-type helices stabilized by intramolecular H bonds between G/C and A/U base pairs. .
![]() Jmol:
Jmol model of tRNA (note the regions of secondary structure - A type
dsRNA) (updated to HTML5 asap)
Jmol:
Jmol model of tRNA (note the regions of secondary structure - A type
dsRNA) (updated to HTML5 asap)
![]() Jmol:
Jmol model
of 70 S Ribosome from T. Thermophilus showing mRNA and tRNA interactions
(updated to HTML5 asap)
Jmol:
Jmol model
of 70 S Ribosome from T. Thermophilus showing mRNA and tRNA interactions
(updated to HTML5 asap)
Problem IB3.1.
The figure below show an actual base pair in a dsDNA.
identify the base pair and the deoxyribose sugars
place a negative charge on any atom with a negative charge
label atoms other than those between the bases that could form H bonds with other DNA binding molecules
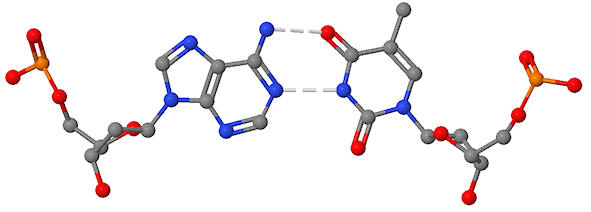
Problem IB3.2.
Do the same for the following base pair
.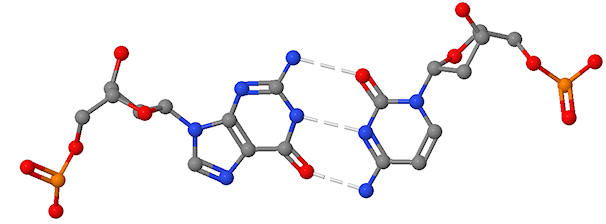
identify the base pair and the deoxyribose sugars
place a negative charge on any atom with a negative charge
label atoms other than those between the bases that could form H bonds with other DNA binding molecules
Problem IB3.3.
The backbone of dsDNA is highlighted below. Place a circle on every atoms with a negative charge.
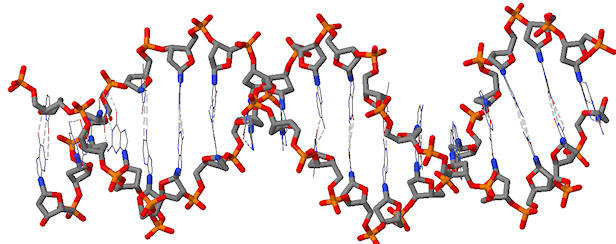
Problem IB3.4.
The structure of a fully protonated peptide, poly-Lys, composed just of the amino acid lysine, is shown below.
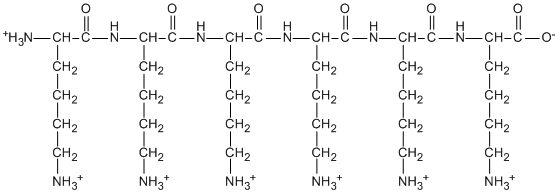
Representing this molecule in cartoon form as an elongated ellipse, show how it might interact with on the DNA helix
Problem IB3.5.
Based on your answer to the above question, what general feature might be present in DNA binding proteins?
This site is written and maintained by Henry Jakubowski as part of a Structure & Reactivity in Organic, Biological and Inorganic Chemistry written by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University. It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry
by
Chris Schaller is licensed under a
Creative
Commons Attribution-NonCommercial 3.0 Unported License.
Send corrections for this chapter to hjakubowski@csbsju.edu
Navigation:
Back to Structure-Property Relationships
Back to Web Materials on Structure & Reactivity in Chemistry