Reactivity in Chemistry
Reduction Potentials of Metal Ions in Biology
MB1. Introduction
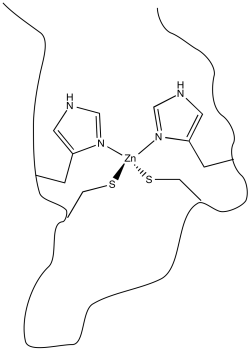
Metal ions play many important roles in biology. Sometimes these roles are structural: a metal ion may simply coordinate to an atom in a biomolecule that acts as a ligand, holding the biomolecule in a particular shape. Zinc finger proteins are a widely studied example. In these cases, a zinc ion binds to the protein in such a way the the protein is held in a particular shape. Generally, this shape serves an important purpose, such as being able to more tightly bind to a DNA molecule or even a small molecule.
In order to bind to the zinc ion, the protein must possess atoms that are able to act as ligands. Lots of atoms in proteins can potentially do so, including, fir instance, the carbonyl oxygen along the protein backbone. However, it is very common for specific amino acid residues to act as ligands. These residues have to be located at just the right position along the protein chain. In the case of zinc finger proteins, one common set of ligands is a pair of histidine residues and a pair of cysteine residues.

Figure MB1.1. A non redox active metal in a biological coordination environment.
In other cases, the metal ion may play the role of a Lewis acid. A substrate that binds to the metal is electrophilically activated; it becomes more able to accept electron donation from a nucleophile. Examples include alcohol dehydrogenases and aldolases, which often contain zinc ions that coordinate carbonyls in the substrate. The activated carbonyl compound undergoes reaction more easily.
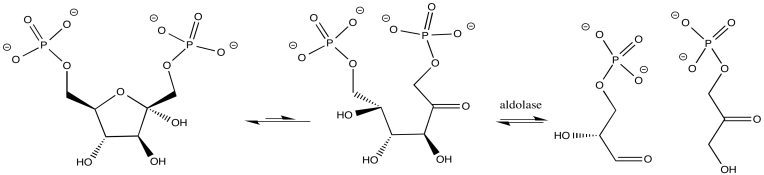
Figure MB1.2. A metal-catalyzed biological reaction.
In many cases, metal ions are redox-active. They may donate or accept an electron from a substrate. Alternatively, they may take part in an electron relay, passing electrons from one to another in order to accomplish electron transport over a long distance.
Ferredoxins are examples of such redox-active sites in biology. Ferredoxins often have a cubic core structure. The corners of the cubes are composed of alternating iron and sulfur atoms. Ferredoxins and related structures are often called iron sulfur clusters.
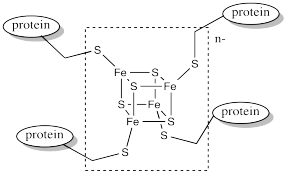
Figure MB1.3. An example of a ferredoxin.
Notice that the iron atoms are held in place via coordination to a surrounding protein. In this case, the irons atoms are bound to cysteine residues. The metal centre is often found buried inside the protein, so that it is completely surrounded by the other amino acid residues. These other resdiues can have a significant impact on the properties of the metal centre, even though they are not directly bound to any metal.
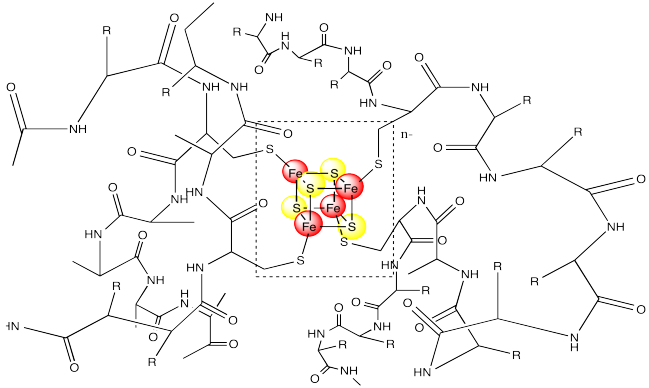
Figure MB1.4. A ferredoxin surrounded by a protein chain.
Problem MB1.1.
The following amino acid residues are the most commonly observed ligands for metal ions in biology. Draw each one at pH 7.
a) cysteine b) histidine c) aspartic acid
Problem MB1.2.
Although the following amino acid residues are not as commonly observed as ligands for metal ions, they are sometimes found in that role. Draw each one at pH 7.
a) methionine b) tyrosine c) glutamic acid d) serine e) threonine
Problem MB1.3.
Lysine is not generally observed to coordinate to metal ions in biology (although there may be exceptions). Explain why, using its structure at pH 7.
The redox potential of the metal ion is vitally important. Reduction potentials must be matched so that the electron transfer will occur easily. The electron transfer may be from the metal to the substrate; in that case, the reduction potential of the substrate must be greater (more positive) than that of the metal ion. Transfer of electron from the metal to the substrate will be favoured.
Of course, if an electron is transferred from the substrate to the metal ion, the opposite would have to be true. The reduction potential of the metal ion would have to be a little more positive than that of the substrate.
In an electron relay, we would need a series of different metal ions, each one with a slightly more positive reduction potential than the last. That way, electrons would keep getting passed from one carrier to the next, and they would travel in the right direction, rather than getting scatterred around randomly.
In order to maintain subtle differences between reduction potentials of metal centres, Nature has developed a wide array of these complexes. Often, the differences between these species involve subtle variations in the coordination environment. For example, a wide variety of iron sulfur clusters are known in biology; a few of them are shown below.
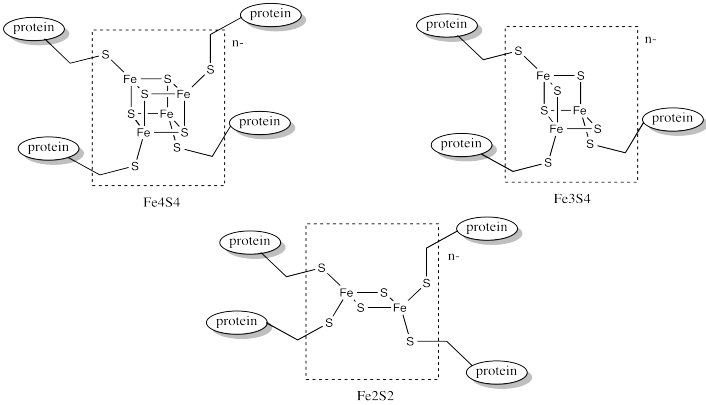
Figure MB1.5. Different examples of iron sulfur (FeS) clusters.
There is a more subtle aspect to these reduction potentials, however. In most cases, there shouldn't be a very large difference between reduction potentials of one species and another.
Why does it matter? Doesn't it seem like a big difference in reduction potentials means the electron transfer is more exothermic? Shouldn't a more exothermic reaction happen even more easily? Isn't that what we want?
Not, really, and there are a couple of reasons why. One reason involves thermodynamic efficiency and heat loss. If a reaction is too exothermic, the energy given off by the reaction is lost as heat energy to the surroundings. Because organisms spend a lot of their time trying to gather energy in order to live, wasting energy isn't a good idea.
The other reason involves the catalytic or cyclic nature of many systems in biology. We can and do re-make lots of chemical species needed for biological activity on a regular basis, but if we can re-use things we don't have to spend as much of our energy re-making and using up reactants over and over again. Lots of species are simply reset to their original state so that they can do their job once more. The easier it is to reset that species, the more often it can keep participating in a cycle, doing the same job over and over again.
In terms of electron transfer, we would like the reduction potential of an electron acceptor to be positive enough so that it easily accepts an electron from its donor. However, we want it to be able to pass its electron along, and get ready to accept another one. That means its reduction potential can't be too positive. It has to be just right.
For reasons like these ones, reduction potentials in biology have to be finely regulated. This problem is as enormously complicated as it is important. In this section, we are going to take a look at a few of the factors that can come into play in modulating reduction potentials in biology. We are certainly not going to gain a great deal of predictive ability, because too many factors can be involved and it will be hard to decide which factor is most important. However, we will be able to understand some of the ways in which different situations influence reduction potentials.
See the section on redox processes in biology at Henry Jakubowski's Biochemistry Online.
This site is written and maintained by Chris P. Schaller, Ph.D., College of Saint Benedict / Saint John's University (retired) with contributions from other authors as noted. It is freely available for educational use.

Structure & Reactivity in Organic, Biological and Inorganic Chemistry by Chris Schaller is licensed under a Creative Commons Attribution-NonCommercial 3.0 Unported License.
This material is based upon work supported by the National Science Foundation under Grant No. 1043566.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Navigation: