Complex IV:
Bovine Cytochrome C
Oxidase
Java version
HTML 5 version (does not require Java; downloads and moves
slowly)
I. Introduction
Complex IV accepts 4 electrons from 4 different reduced cytochrome C hemes
and transfers them to dioxygen to produce 2 H2O molecucles, Dioxygen
gets reduced. In the process 4H+s are added in forming the
water, and 4H+s are vectorially moved from the matrix to the inner
membrane space. .
For more information see
Biochemistry Online: Chapter 8C-7 - Complex IV
II. General Structure
A. Electron transfer
Electrons flow from reduced cytochrome C to a dinuclear Cu cluster (CuA) to an adjacent heme a (low spin) which transfers them to another heme a3 (high spin) and then finally to dioxygen which is coordinated to the Fe in heme a3 and to an adjacent CuB. The heme a3 Fe:Cu dinuclear cluster is unique among all hemes.
Cartoon of Complex IV without electron carriers
Now let's sequentially add the bound electron carriers which receive electrons from the mobile
and soluble reduced cytochrome C delivers them to the mobile
soluble acceptor, dioxygen.
add the dinuclear CuA cluster
add the adjacent low spin heme a
add the high spin heme a3, which is adjacent to a nearby Cu
add the CuB ion which helpd coordinate dioxygen and its reduction products to heme a3
add peroxide which is found in this pdb file. It forms a ligand bridge between the CuB and heme a3 and helps keep reduction products of dioxygen from dissociating from the complex before full reduction to water.
view all bound electron carriers without the protein
B. Proton Transfer
Proton transfer is as critical as electron transfer as it is necessary to
created the protonmotive force which powers ATP synthesis by the FoF1 ATP
synthase.
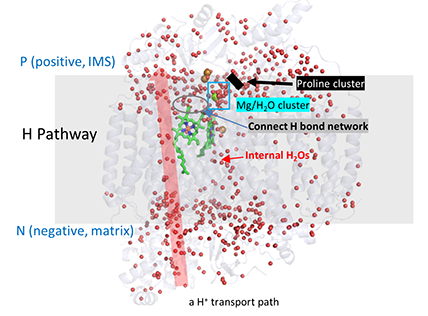
Proton transfer involves a water channel in the protein and series of connected
H bond donors and acceptors. The water channel runs from the N
(negative) side ( matrix side), where water or hydronium ion can enter the
channel, to the start of the internal H bond network, which then extends to
the P (positive) side (inner membrane space). This is called the H pathway.
It is connected through a small interconnecting set of H bond donors and
acceptor to a Mg ion/H2O cluster. This site accumulates
protons for eventual reduction of water. The Mg/water cluster can exchange its water
with the IMS space but that exchange is potentially blocked by a Prolyl cluster.
These features are depicted in the figure below (which shows internal water
molecules as red dots) and in the modeling buttons below.
Add Mg ion
Add internal water molecules (determined by Pymol). The membrane plane runs through the parallel alpha helices.
Show waters listed in Yano JBC paper in the Mg ion/water cluster (oxidized complex).
Add proline cluster which stops exchange of water with Mg/water cluster.
H pathway for H+ transport and connection to Mg ion water cluster.
The light blue spheres are the water cluster surrounding the Mg ion. The arrow (from the negative N side to the positive P side shows the H bond donors/acceptors
in the H pathway. The water cluster is connected to the H pathway by H bonds from R439 and Y371 (wireframe) and also proprionates (labeled) on heme a.