Biochemistry Online: An Approach Based on Chemical Logic

CHAPTER 8 - OXIDATION/PHOSPHORYLATION
B: OXIDATIVE ENZYMES
BIOCHEMISTRY - DR. JAKUBOWSKI
04/15/16
|
Learning Goals/Objectives for Chapter 8B: After class and this reading, students will be able to
|
B3. DEHYDROGENASES
These enzymes usually involve NAD+/NADH and are named for the substrate
that is oxidized by NAD+. For instance in the reaction:
pyruvate + NADH <===> lactate + NAD+
which is used to regenerate NAD+ under anerobic conditions, the enzyme is named lactate dehydrogenase. As in acid/base reactions, when the preferred direction for the reaction (from a ΔGo perspective) is from stronger acid to weaker (conjugate) acid, the preferred direction for a redox reaction is in the direction from strong to weak oxidizing/reducing agents. This can easily be determined from charts of standard reduction potentials, and using the equation: ΔGo = -nFΔEo,
- where F is the Faraday constant (96,494 Coulombs/mol e- = 96, 494 J/(V.mol) = 23.06 kcal/(V.mol) . One Faraday is the charge per one mol of electrons).
- andΔEo, the standard EMF or standard cell potential (total voltage at standard state conditions), which can be determined by adding the standard reduction potentials (Eo) for the two appropriate half-reactions, after reversing the equation for the half-reaction that represents the oxidation.
When n=2 (number of electrons) which is common for oxidations of organic molecules,
ΔGo (kcal/mol) = - 46.12ΔEo or for government work ΔGo (kcal/mol) = - 50ΔEo
Notice when ΔEo > 0, ΔGo < 0, the reaction as written is favored under standard conditions. Note in the table below that many of the half reactions involve protons. For biological reactions involving free protons, the standard state concentration for the protons are not 1 M as for other solutes in solution, but defined to be the hydronium ion concentration at pH 7.0. The ΔEo and ΔGo values for the reactions involving hydrogen ions at a standard state of pH 7.0 are usually written as ΔEo' and ΔGo'
Table: Standard Reduction Potential Table (E0'), 25oC
| oxidant |
reductant |
n (electrons) | Eo� (volts) |
| Acetate + carbon dioxide |
pyruvate |
2 | -0.70 |
| succinate + CO2 + 2H+ |
α−ketoglutarate + H2O |
2 | -0.67 |
| acetate |
acetaldehyde |
2 | -0.60 |
| glycerate-3-P | glyceraldehyde-3-P + H2O | 2 | -0.55 |
| O2 | O2- | 1 | -0.45 |
| ferredoxin (ox) | ferredoxin (red) | 1 | -0.43 |
| Carbon dioxide |
formate |
2 | -0.42 |
| 2H+ |
H2 |
2 | -0.42 |
| α-ketoglutarate + CO2 + 2H+ |
isocitrate |
2 | -0.38 |
| acetoacetate | β-hydroxybutyrate | 2 | -0.35 |
| Cystine |
cysteine |
2 | -0.34 |
| Pyruvate + CO2 |
malate |
2 | -0.33 |
| NAD+ + 2H+ |
NADH + H+ |
2 | -0.32 |
| NADP+ + 2H+ |
NADPH + H+ |
2 | -0.32 |
| FMN (enzyme bound) | FMNH2 | 2 | -0.30 |
| Lipoic acid, ox |
Lipoic acid, red |
2 | -0.29 |
| 1,3 bisphosphoglycerate + 2H+ |
glyceraldehyde-3-P + Pi |
2 | -0.29 |
| Glutathione, ox |
red |
2 | -0.23 |
| FAD (free) + 2H+ |
FADH2 |
2 | -0.22 |
| Acetaldehyde + 2H+ |
ethanol |
2 | -0.20 |
| Pyruvate + 2H+ |
lactate |
2 | -0.19 |
| Oxalacetate + 2H+ |
malate |
2 | -0.17 |
| α-ketoglutarate + NH4+ |
glutamate |
2 | -0.14 |
| FAD + 2H+ (bound) |
FADH2 (bound) |
2 | 0.003-0.09 |
| Methylene blue, ox |
Methylene blue, red |
2 | 0.01 |
| Fumarate + 2H+ |
succinate |
2 | 0.03 |
| CoQ (Ubiquinone - UQ + H+ | UQH. | 1 | 0.031 |
| UQ + 2H+ | UQH2 | 2 | 0.06 |
| Dehydroascorbic acid |
ascorbic acid |
2 | 0.06 |
| Ubiquinone; ox |
red |
2 | 0.10 |
| Cytochrome b2; Fe3+ |
Cytochrome b2; Fe2+ |
1 | 0.12 |
| Cytochrome c1; Fe3+ |
Cytochrome c1; Fe2+ |
1 | 0.22 |
| Cytochrome c; Fe3+ |
Cytochrome c; Fe2+ |
1 | 0.25 |
| Cytochrome a; Fe3+ |
Cytochrome a; Fe2+ |
1 | 0.29 |
| 1/2 O2 + H2O |
H2O2 |
2 | 0.30 |
| Cytochrome a3; Fe3+ |
Cytochrome a3; Fe2+ |
1 | 0.35 |
| Ferricyanide |
ferrocyanide |
2 | 0.36 |
| Cytochrome f; Fe3+ |
Cytochrome f; Fe2+ |
1 | 0.37 |
| Nitrate |
nitrite |
1 | 0.42 |
| Photosystem P700 | . | . | 0.43 |
| Fe3+ |
Fe2+ |
1 | 0.77 |
| 1/2 O2 + 2H+ |
H2O |
2 | 0.816 |
-
 Recommendations
for nomenclature and tables in biochemical thermodynamics (from the
International Union of Biochemistry and Molecular Biology and the IUPAC)
Recommendations
for nomenclature and tables in biochemical thermodynamics (from the
International Union of Biochemistry and Molecular Biology and the IUPAC)
The mechanism for the oxidation of a substrate by NAD + involves concerted hydride transfer to one face of NAD+.
Consider for example the oxidation of ethanol to acetaldehyde by alcohol dehydrogenase.
Figure: oxidation of ethanol to acetaldehyde by alcohol dehydrogenase
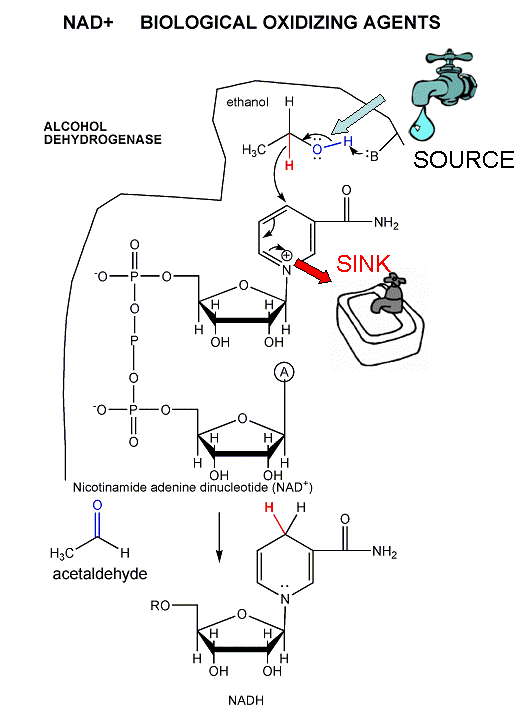
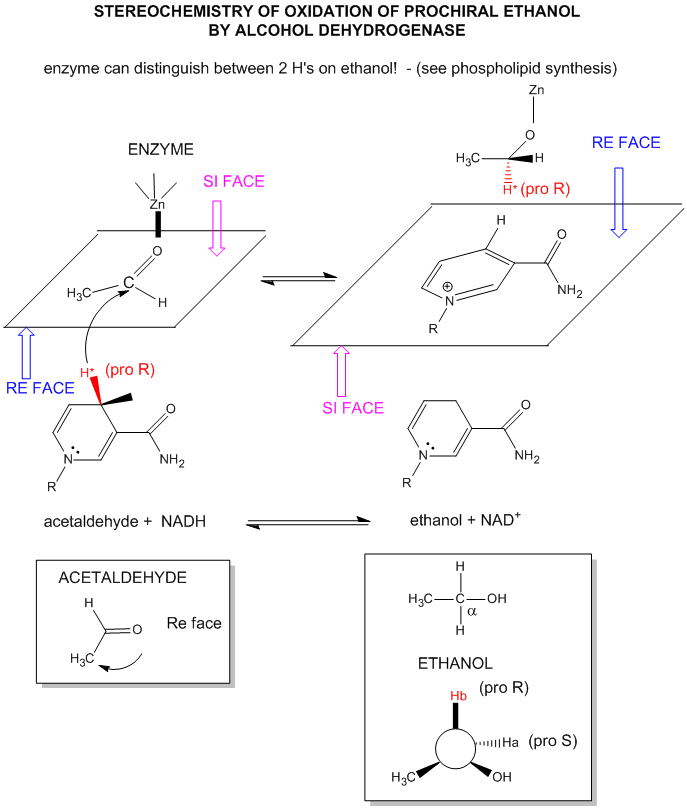
For substrates like ethanol that loses a hydride from a methylene carbon atom that has two H's, only one of the H's is lost (either the proR or proS) from the prochiral center. (Remember the reaction of prochiral glycerol to give phospholipids.)
Figure: STEREOCHEMISTRY OF NAD+/NADH REDOX REACTIONS WITH ALCOHOL DEHYDROGENASE
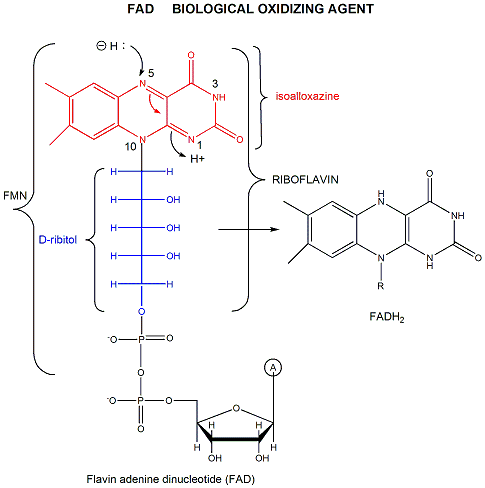
FAD has a more positive reduction potential than NAD+ so it is used for more "demanding" oxidation reactions, such as dehydrogenation of a C-C bond to form an alkene. You will notice on standard reduction potential tables that the potential of FAD is often listed several times and depends on the enzyme. This is because the FAD is tightly bound to the enzyme so its tendency to acquire electrons depends on its environment, in much the same fashion as the pKa of an amino acid side chain (which reflects is tendency to release protons) is affected by the environment of the amino acid side chain in the protein. The standard reduction potential for flavin enzymes varies from -465 mV to + 149 mV. Compare this to the reduction potential of free FAD/FADH2, which in aqueous solution is -208 mV. The standard reduction potential of the flavin in D-amino acid oxidase, a flavoprotein, is about 0.0 V. Remember, the more positive the standard reduction potential, the more likely the reactant will be reduced and hence act as an oxidizing agent. Hence the FAD in D-amino acid oxidase is a better oxidizing agent than free FAD. The Kd for binding of FAD to the enzyme is 10-7M compared to the Kd for binding of FADH2, which is 10-14M. By gaining electrons, the flavin binds more tightly, which preferentially stabilizes the bound FADH2 compared to the bound FAD. This shifts the equilibrium of FAD <=> FADH2 to the right, making the bound FAD a stronger oxidizing agent.
Figure: FAD AND OXIDATIONS: MECHANISM
![]() Jmol:
Updated Flavin dehydrogenase
Jmol14 (Java) |
JSMol (HTML5)
Jmol:
Updated Flavin dehydrogenase
Jmol14 (Java) |
JSMol (HTML5)
Can the standard reduction potential of a redox active center in a protein be tuned by changing the environment of that center, much as the pKa of an acid side chain can by changing the polarity of the environment? The answer is yes. The active site of azurin, a cupredoxin, has a redox active copper ion coordinated by a Cys and two His residues in a trigonal planar fashion. Met 121 serves as a weak axial ligand. Marshall et al. have reported a feasible method to manipulate the redox potential (Eo) of this active site. The wild type azurin was mutated to alter the hydrophobicity and hydrogen bonding capabilities, while maintaining the overall architecture of the metal binding site. Ser 46 was selected for mutation since it occupied a position similar to Asn in another curedoxin that was involved in an important H bond binding two ligand binding loops. An N47S-mutation, which strengthened the hydrogen bond between the two ligand-containing loops increased Eo by ~130 mV while preserving metal binding site architecture as determined by UV-Vis spectroscopy. They also compared a M121Q mutant with wild-type M121 and with a M121L mutant. A plot of Eo vs log partition coefficient for transfer of the side chain from water to octanol was essentially linear with a positive slope, showing that the standard reduction potential depended on the hydrophobicity of the weakly coordinating ligand in the metal binding region. This behavior extended to double mutants (where one set of mutants involved M121). The investigators were able to tune the Eo over a 700 mv range!
.
Navigation
Return to Chapter 8B: Oxidative Enzymes Sections
Return to Biochemistry Online Table of Contents
Archived version of full Chapter 8B: Oxidative Enzymes

Biochemistry Online by Henry Jakubowski is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.